-
PDF
- Split View
-
Views
-
Cite
Cite
Paula Tamagnini, Elsa Leitão, Paulo Oliveira, Daniela Ferreira, Filipe Pinto, David James Harris, Thorsten Heidorn, Peter Lindblad, Cyanobacterial hydrogenases: diversity, regulation and applications, FEMS Microbiology Reviews, Volume 31, Issue 6, November 2007, Pages 692–720, https://doi.org/10.1111/j.1574-6976.2007.00085.x
Close - Share Icon Share
Abstract
Cyanobacteria may possess two distinct nickel-iron (NiFe)-hydrogenases: an uptake enzyme found in N 2 -fixing strains, and a bidirectional one present in both non-N 2 -fixing and N 2 -fixing strains. The uptake hydrogenase (encoded by hupSL ) catalyzes the consumption of the H 2 produced during N 2 fixation, while the bidirectional enzyme ( hoxEFUYH ) probably plays a role in fermentation and/or acts as an electron valve during photosynthesis. hupSL constitute a transcriptional unit, and are essentially transcribed under N 2 -fixing conditions. The bidirectional hydrogenase consists of a hydrogenase and a diaphorase part, and the corresponding five hox genes are not always clustered or cotranscribed. The biosynthesis/maturation of NiFe-hydrogenases is highly complex, requiring several core proteins. In cyanobacteria, the genes that are thought to affect hydrogenases pleiotropically ( hyp ), as well as the genes presumably encoding the hydrogenase-specific endopeptidases ( hupW and hoxW ) have been identified and characterized. Furthermore, NtcA and LexA have been implicated in the transcriptional regulation of the uptake and the bidirectional enzyme respectively. Recently, the phylogenetic origin of cyanobacterial and algal hydrogenases was analyzed, and it was proposed that the current distribution in cyanobacteria reflects a differential loss of genes according to their ecological needs or constraints. In addition, the possibilities and challenges of cyanobacterial-based H 2 production are addressed.
Introduction
Cyanobacteria, one of the largest and most important groups of bacteria on Earth, are able to perform oxygenic photosynthesis using water as an electron donor and may be found in almost any ecological niche from fresh to salt water, terrestrial and extreme environments ( Whitton & Potts, 2000 ). The knowledge on such a diverse group of prokaryotic organisms has greatly increased since cyanobacterial genomes became available. In 1996, the entire sequence of Synechocystis sp. PCC 6803 was published ( Kaneko et al. , 1996 ; Nakamura et al. , 1998 ), and since then, many other cyanobacterial genome projects have been completed and released, including that of Nostoc punctiforme ATCC 29133/PCC 73102, one of the largest microbial genomes sequenced so far ( Meeks et al. , 2001 ; Anderson et al. , 2006 ).
Fossil traces of cyanobacteria are claimed to have been found from around 3.5 billion years ago ( Schopf, 2000 ), and ancestors of cyanobacteria most probably played a key role in the formation of atmospheric oxygen, and are thought to have evolved into present-day chloroplasts of algae and green plants ( Miyagishima, 2005 ; Mulkidjanian et al. , 2006 ). Cyanobacteria display a relatively wide range of morphological diversity, including unicellular, filamentous and colonial forms. Some filamentous strains form differentiated cells specialized in nitrogen fixation – heterocysts, and spore-like resting cells – akinetes. A number of nonheterocystous strains are also able to perform N 2 fixation under certain conditions. The fact that several cyanobacteria are able to reduce nitrogen and carbon under aerobic conditions may be responsible for their evolutionary and ecological success. In cyanobacteria, as in any diazotrophic bacteria, the reduction of N 2 to NH 3 is accompanied by the formation of molecular hydrogen ( Berman-Frank et al. , 2003 ). The H 2 produced by the nitrogenase is rapidly consumed by an uptake hydrogenase, an enzyme that has been found in almost all the N 2 -fixing cyanobacteria examined so far, with one reported exception – Synechococcus sp. BG 043511 ( Ludwig et al. , 2006 ). Additionally, these strains may contain a bidirectional hydrogenase, an enzyme that is generally present in the non nitrogen-fixing cyanobacteria ( Tamagnini et al. , 2002 , 2005 ), but absent in Gloeobacter violaceus PCC 7421, a cyanobacterium that possesses a number of unique characteristics such as the absence of thylakoids ( Nakamura et al. , 2003 ; Ludwig et al. , 2006 ). The distribution of genes related to hydrogenases among representative cyanobacterial strains is displayed in Table 1 . Both cyanobacterial hydrogenases are NiFe enzymes, which are the most common hydrogenases found in bacteria and Archaea . The core enzyme consists of an αβ heterodimer with the large/α subunit hosting the bimetallic active site, and the small/β-subunit containing the FeS clusters, which function as electron transfer domains between the electron acceptors/donors and the catalytic center of the enzyme ( Fig. 1 ). In general, the NiFe hydrogenases are divided into four groups, with the cyanobacterial uptake hydrogenases clustering together with the cytoplasmic H 2 sensors of group 2, and the bidirectional enzymes belonging to group 3 comprising the bidirectional heteromultimeric cytoplasmic hydrogenases (for reviews on this subject, see Vignais et al. , 2001 ; Vignais & Colbeau, 2004 ).
Distribution of genes related to hydrogenases in representative cyanobacterial strains
| Organisms | Bidirectional hydrogenase | Uptake hydrogenase | hupL recombinase | Bidirectional specific endopeptidase | Uptake specific endopeptidase | Other maturation genes | GenBank accession number/ References | |||
| hoxFUYH | hoxE | hupSL | XisC* | hoxW | hupW | hypFCDEAB | ||||
| Unicellular non-N 2 -fixing | G. violaceus PCC 7421 | − | − | − | − | − | − | NC_005125Nakamura, (2003) | ||
| Synechocystis sp. PCC 6803 | + Appel & Schulz (1996) | + | − | + | − | + Scattered | NC_000911Kaneko, (1996) | |||
| Unicellular N 2 -fixing | C. watsonii WH 8501 | − | − | + | − | + | + Scattered | NZ_ADV00000000 | ||
| Filamentous nonheterocystous | L. majuscula CCAP 1446/4 | + | ND | + | ND | + | + Operon | Leitão, (2005 , 2006) | ||
| N 2 -fixing | T. erythraeum IMS 101 | − | − | + | − | + | + | NC_008312 | ||
| Filamentous heterocystous N 2 -fixing | A. variabilis ATCC 29413 | + Schmitz, (1995) | + | + Happe, (2000) | − | + | + | + | NC_007413 | |
| Nostoc sp. PCC 7120 | + | + | + Carrasco, (1995) | + | + | + | + Gubili & Borthakur (1996 , 1998) | NC_003272Kaneko, (2001) | ||
| N. punctiforme PCC 73102 | − | − | + Oxelfelt, (1998) | − | − | + | + Operon Hansel, (2001) | NZ_AAAY00000000 | ||
| Organisms | Bidirectional hydrogenase | Uptake hydrogenase | hupL recombinase | Bidirectional specific endopeptidase | Uptake specific endopeptidase | Other maturation genes | GenBank accession number/ References | |||
| hoxFUYH | hoxE | hupSL | XisC* | hoxW | hupW | hypFCDEAB | ||||
| Unicellular non-N 2 -fixing | G. violaceus PCC 7421 | − | − | − | − | − | − | NC_005125Nakamura, (2003) | ||
| Synechocystis sp. PCC 6803 | + Appel & Schulz (1996) | + | − | + | − | + Scattered | NC_000911Kaneko, (1996) | |||
| Unicellular N 2 -fixing | C. watsonii WH 8501 | − | − | + | − | + | + Scattered | NZ_ADV00000000 | ||
| Filamentous nonheterocystous | L. majuscula CCAP 1446/4 | + | ND | + | ND | + | + Operon | Leitão, (2005 , 2006) | ||
| N 2 -fixing | T. erythraeum IMS 101 | − | − | + | − | + | + | NC_008312 | ||
| Filamentous heterocystous N 2 -fixing | A. variabilis ATCC 29413 | + Schmitz, (1995) | + | + Happe, (2000) | − | + | + | + | NC_007413 | |
| Nostoc sp. PCC 7120 | + | + | + Carrasco, (1995) | + | + | + | + Gubili & Borthakur (1996 , 1998) | NC_003272Kaneko, (2001) | ||
| N. punctiforme PCC 73102 | − | − | + Oxelfelt, (1998) | − | − | + | + Operon Hansel, (2001) | NZ_AAAY00000000 | ||
Rearragement occurring during the differentiation of a vegetative cell into a heterocyst.
ND, not determined.
Distribution of genes related to hydrogenases in representative cyanobacterial strains
| Organisms | Bidirectional hydrogenase | Uptake hydrogenase | hupL recombinase | Bidirectional specific endopeptidase | Uptake specific endopeptidase | Other maturation genes | GenBank accession number/ References | |||
| hoxFUYH | hoxE | hupSL | XisC* | hoxW | hupW | hypFCDEAB | ||||
| Unicellular non-N 2 -fixing | G. violaceus PCC 7421 | − | − | − | − | − | − | NC_005125Nakamura, (2003) | ||
| Synechocystis sp. PCC 6803 | + Appel & Schulz (1996) | + | − | + | − | + Scattered | NC_000911Kaneko, (1996) | |||
| Unicellular N 2 -fixing | C. watsonii WH 8501 | − | − | + | − | + | + Scattered | NZ_ADV00000000 | ||
| Filamentous nonheterocystous | L. majuscula CCAP 1446/4 | + | ND | + | ND | + | + Operon | Leitão, (2005 , 2006) | ||
| N 2 -fixing | T. erythraeum IMS 101 | − | − | + | − | + | + | NC_008312 | ||
| Filamentous heterocystous N 2 -fixing | A. variabilis ATCC 29413 | + Schmitz, (1995) | + | + Happe, (2000) | − | + | + | + | NC_007413 | |
| Nostoc sp. PCC 7120 | + | + | + Carrasco, (1995) | + | + | + | + Gubili & Borthakur (1996 , 1998) | NC_003272Kaneko, (2001) | ||
| N. punctiforme PCC 73102 | − | − | + Oxelfelt, (1998) | − | − | + | + Operon Hansel, (2001) | NZ_AAAY00000000 | ||
| Organisms | Bidirectional hydrogenase | Uptake hydrogenase | hupL recombinase | Bidirectional specific endopeptidase | Uptake specific endopeptidase | Other maturation genes | GenBank accession number/ References | |||
| hoxFUYH | hoxE | hupSL | XisC* | hoxW | hupW | hypFCDEAB | ||||
| Unicellular non-N 2 -fixing | G. violaceus PCC 7421 | − | − | − | − | − | − | NC_005125Nakamura, (2003) | ||
| Synechocystis sp. PCC 6803 | + Appel & Schulz (1996) | + | − | + | − | + Scattered | NC_000911Kaneko, (1996) | |||
| Unicellular N 2 -fixing | C. watsonii WH 8501 | − | − | + | − | + | + Scattered | NZ_ADV00000000 | ||
| Filamentous nonheterocystous | L. majuscula CCAP 1446/4 | + | ND | + | ND | + | + Operon | Leitão, (2005 , 2006) | ||
| N 2 -fixing | T. erythraeum IMS 101 | − | − | + | − | + | + | NC_008312 | ||
| Filamentous heterocystous N 2 -fixing | A. variabilis ATCC 29413 | + Schmitz, (1995) | + | + Happe, (2000) | − | + | + | + | NC_007413 | |
| Nostoc sp. PCC 7120 | + | + | + Carrasco, (1995) | + | + | + | + Gubili & Borthakur (1996 , 1998) | NC_003272Kaneko, (2001) | ||
| N. punctiforme PCC 73102 | − | − | + Oxelfelt, (1998) | − | − | + | + Operon Hansel, (2001) | NZ_AAAY00000000 | ||
Rearragement occurring during the differentiation of a vegetative cell into a heterocyst.
ND, not determined.
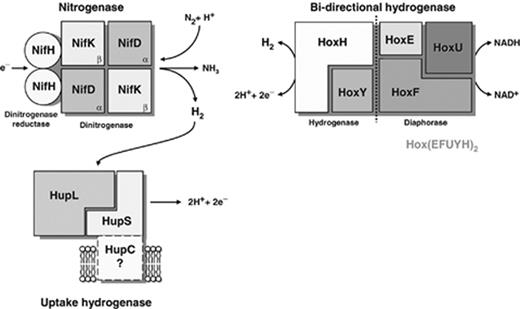
Enzymes directly involved in hydrogen metabolism in cyanobacteria. While the uptake hydrogenase is present in most of the nitrogen-fixing strains tested (with only one exception reported so far; see text and Table 1 ), the bidirectional enzyme seems to be present in non-N 2 -fixing and N 2 -fixing strains but is not a universal enzyme. The existence of a third subunit (HupC) anchoring the uptake hydrogenase to the membrane is yet to be confirmed, and the molecular weight of the native bidirectional hydrogenase indicates a dimeric assembly of the enzyme complex Hox(EFUYH) 2 .
In the present review, recent advances on cyanobacterial hydrogenases, have been summarized focusing on achievements on the diversity and molecular regulation of both the uptake and the bidirectional enzyme.
Photobiological production of H 2 by microorganisms is of great public interest because it promises a renewable energy carrier from nature's most plentiful resources: solar energy and water. Cyanobacteria and green algae are the only organisms known so far that are capable of both oxygenic photosynthesis and hydrogen production. In a separate section, the possibilities and challenges in cyanobacterial-based hydrogen production are outlined.
Uptake hydrogenase
The cyanobacterial uptake hydrogenase, found exclusively in N 2 -fixing strains and encoded by the hup – hydrogen uptake – genes, is at least a heterodimeric enzyme with a large subunit of about 60 kDa containing the active site (HupL) and a small subunit of c . 35 kDa playing a role in electron transfer (HupS) ( Fig. 1 ). Because the physiological and biochemical data point to a membrane-bound enzyme ( Houchins & Burris, 1981b ; Houchins et al. , 1984 ; Lindblad & Sellstedt, 1990 ; Rai et al. , 1992 ), and the hydropathy profiles of the HupL and the HupS proteins do not indicate any transmembrane domains ( Tamagnini et al. , 2005 ), the existence of a polypeptide that anchors the HupSL heterodimer to the membrane seems likely. In fact, analysis of the available genomes revealed the presence of ORFs whose products could potentially fulfill this anchoring role ( Lindberg et al. , 2003 ). However, to date no definitive proof was obtained, and the existence of both a soluble and a membrane-bound form of the enzyme cannot be excluded (see for e.g. Houchins & Burris, 1981b ).
Immunolocalization studies, using antibodies produced against hydrogenases from other bacteria, showed that the hydrogenase antigens are present in both the vegetative cells and heterocysts of N. punctiforme , and several symbiotic Nostoc strains ( Lindblad & Sellstedt, 1990 ; Rai et al. , 1992 ; Tamagnini et al. , 1995 ). However, these studies do not clarify whether the enzyme is in its active form in both cell types. In Anabaena / Nostoc sp. PCC 7120, the uptake hydrogenase activity was essentially associated with the particulate fraction of the heterocysts ( Houchins & Burris, 1981b ); however, one must bear in mind that in this strain the hupL gene undergoes a rearrangement, allowing its expression in the heterocysts only, and that this process does not occur in N. punctiforme ( Oxelfelt et al. , 1998 ). Moreover, the presence/levels of the cyanobacterial uptake hydrogenase are certainly dependent on the growth conditions. In heterocystous cyanobacteria grown in air and without combined nitrogen, the uptake hydrogenase activity is mainly confined to heterocysts, where it is protected from oxygen inactivation; however, the exact location of the enzyme in cyanobacteria should be further investigated in both heterocystous and nonheterocystous strains.
A strong correlation between the nitrogen-fixation process and the uptake hydrogenase activity has been demonstrated for cyanobacteria ( Lambert & Smith, 1981 ; Houchins et al. , 1984 ; Wolk et al. , 1994 ; Oxelfelt et al. , 1995 ; Schütz, 2004 ), and this indicates that the main physiological function of the uptake hydrogenase is to reutilize and regain the H 2 /electrons produced by the H 2 evolution through the nitrogenase. This recycling has been suggested to have at least three beneficial functions to the organism: (1) it provides ATP via the oxyhydrogen reaction, minimizing the loss of energy; (2) it removes the oxygen from nitrogenase, thereby protecting it from inactivation; and (3) it supplies reducing equivalents (electrons) to various cell functions ( Bothe et al. , 1977 , 1991 ; Howarth & Codd, 1985 ; Weisshaar & Böger, 1985 ; Smith et al. , 1990 ).
Physical organization of hup genes and the corresponding proteins
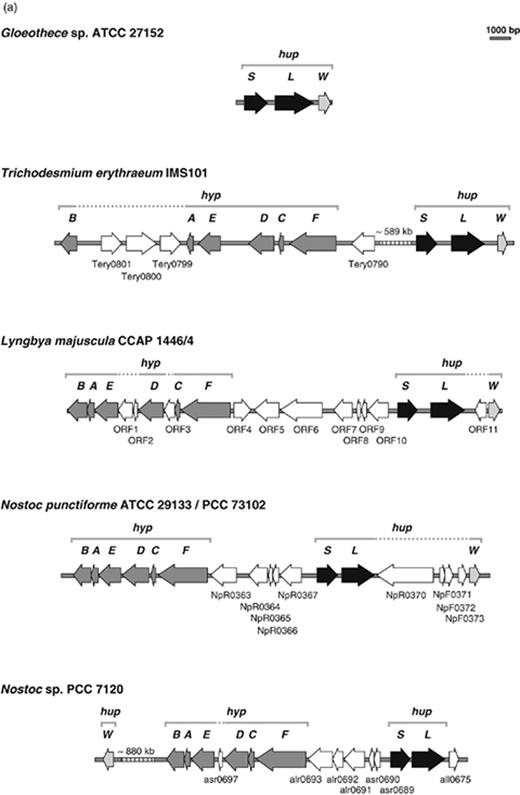
The physical arrangement of the structural genes encoding the uptake hydrogenase is very similar in all the cyanobacteria studied so far: hupS and hupL are contiguous, with the gene encoding the smaller subunit located upstream from the gene encoding the larger one ( Carrasco et al. , 1995 ; Oxelfelt et al. , 1998 ; Happe et al. , 2000 ; Lindberg et al. , 2000 ; Oliveira et al. , 2004 ; Leitão, 2005 ) ( Fig. 2 ). Transcriptional start sites have been identified upstream of the hupS start codon ( Happe et al. , 2000 ; Lindberg et al. , 2000 ; Oliveira et al. , 2004 ; Leitão, 2005 ), and a putative transcriptional terminator, located immediately downstream of hupL , has been found in N. punctiforme ( Lindberg et al. , 2000 ). In agreement, reverse transcriptase (RT)-PCR experiments, and the sizes of transcripts determined by Northern blot, indicate that hupSL constitute a transcriptional unit in Anabaena variabilis ATCC 29413, N. punctiforme and Lyngbya majuscula CCAP 1446/4 ( Happe et al. , 2000 ; Lindberg et al. , 2000 ; Leitão, 2005 ). In the unicellular Gloeothece sp. ATCC 27152 and in the filamentous Trichodesmium erythraeum IMS 101 hupW – the gene encoding for the putative uptake hydrogenase-specific endopeptidase – is the ORF located immediately downstream of hupL , and was shown to be cotranscribed with hupSL in Gloeothece sp. ATCC 27152 ( Oliveira et al. , 2004 ). In other strains, the position of hupW related to the hupSL varies considerably, and in the strains examined they are transcribed independently ( Wünschiers, 2003 ) ( Fig. 2 ).
Organization of the loci containing the genes encoding (a) the uptake hydrogenase ( hup ) and (b) the bidirectional hydrogenase ( hox ) in selected cyanobacterial strains (black ORFs). The accessory genes ( hyp , hupW and hoxW ), encoding proteins involved in the maturation of the hydrogenases are also depicted, as gray ORFs, as well as some additional ORFs (identified, when available, with the corresponding ORF-number in respective annotated genomes, and shown as white ORFs). Gloeothece sp. ATCC27152 ( Oliveira et al. , 2004 – GenBank accession no. AY260103 ), Trichodesmium erythraeum IMS101 ( http://genome.jgi-psf.org/finished_microbes/trier/trier.home.html ), Lyngbya majuscula CCAP 1446/4 ( Leitão et al. , 2005 – GenBank accession no. AF368526 ), Nostoc punctiforme ATCC 29133/PCC 73102 ( http://genome.jgi-psf.org/draft_microbes/nospu/nospu.home.html ), Nostoc sp. PCC 7120 ( Kaneko et al. , 2001 ), Synechocystis sp. PCC 6803 ( Kaneko et al. , 1996 ), Synechococcus elongatus PCC 7942 ( http://genome.jgi-psf.org/finished_microbes/synel/synel.home.html ), Arthrospira platensis FACHB341 ( Zhang et al. , 2005a , b – GenBank accession nos. DQ309870 and AY345594 ) and Anabaena variabilis ATCC 29413 ( http://genome.jgi-psf.org/finished_microbes/anava/anava.home.html ).
Analysis of the predicted proteins encoded by the hupSL operon demonstrated that whereas HupS has the same number of amino acid residues in all the cyanobacteria investigated [320 amino acids (aa)], HupL generally has 531 aa with the exception of the filamentous nonheterocystous L. aestuarii CCY 9616 (six extra), L. majuscula (six extra), and T. erythraeum (three extra). To date, the physiological significance (if any) of these extra residues is still unknown.
In the NiFe hydrogenases, the large subunit harbors the active center that is deeply buried inside the protein, close to the large interface between the two subunits, and the small subunit contains the FeS clusters that conduct electrons between the active center and the physiological electron acceptor/donor ( Vignais et al. , 2001 ; Vignais & Colbeau, 2004 ). In concordance, the cyanobacterial HupL sequences contain the four conserved cysteine residues that are involved in the coordination of the bimetallic NiFe center of the active site, and HupS contains eight cysteine residues clearly corresponding to those involved in the formation of the FeS clusters, and a ninth cysteine slightly shifted compared with other bacteria ( Tamagnini et al. , 2002 ). In addition, HupL contains the C-terminal region that is presumably cleaved off, by a specific endopeptidase, as the last step of the maturation of the large subunit. In contrast with other organisms, HupS lacks both the twin-arginine signal peptide at the N-terminal, and the hydrophobic motif at the C-terminal proposed to be involved in translocation and anchorage to the membrane, respectively. As mentioned previously, these general features of the cyanobacterial hydrogenases cluster them together with the soluble H 2 -sensing enzymes ( Vignais et al. , 2001 ; Vignais & Colbeau, 2004 ). However, the construction of hup− mutants proved that the cyanobacterial uptake hydrogenase is indeed a physiological functional enzyme rather than a regulatory one ( Happe et al. , 2000 ; Lindberg et al. , 2002 ; Lindblad et al. , 2002 ; Masukawa et al. , 2002 ).
hupL rearrangement in heterocystous strains
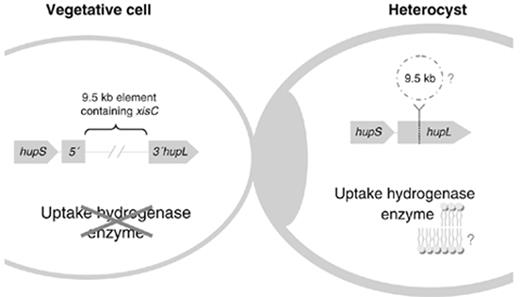
Programmed DNA rearrangements have been described in eukaryotes and prokaryotes but are relatively uncommon events. In cyanobacteria, developmentally regulated DNA rearrangements have been reported to occur in heterocystous strains (for a review, see Golden et al. , 1997 ). Generally, the ORF is interrupted in the vegetative cells by a 10–60-kb DNA element, which is excised during the differentiation of a photosynthetic vegetative cell into a N 2 -fixing heterocyst, restoring the structure of the gene/operon and allowing its expression in heterocysts only.
The rearrangement within hupL (large subunit of the uptake hydrogenase) was first described for Nostoc sp. PCC 7120 ( Carrasco et al. , 1995 ). In the vegetative cells of this cyanobacterium, hupL is interrupted by a 9.5-kb element that is excised late during the heterocyst differentiation process by a site-specific recombination between the 16-bp direct repeats that flank the element ( Fig. 3 ). The hupL element contains, in one of its borders, the gene that encodes the recombinase necessary for the excision – xisC ( Carrasco et al. , 1995 , 1998 , 2005 ). Site-directed mutagenesis revealed that the XisC protein has a functional similarity to the phage integrase family of recombinases. Recently, it has been unequivocally demonstrated that the inactivation of xisC blocks the hupL rearrangement and that XisC alone is sufficient to catalyze the hupL element site-specific recombination in Nosto c sp. PCC 7120 ( Carrasco et al. , 2005 ). It was also shown that the xisC -mutant forms heterocysts without any obvious developmental defects and that the mutant grown under N 2 -fixing conditions (BG11 0 ) was not only defective for hydrogen uptake activity but evolves H 2 ( Lindblad et al. , 2002 ; Carrasco et al. , 2005 ). Moreover, Lindblad, (2002) showed that, in a competitive growth environment with increased light intensity, the wild-type strain has an advantage over the xisC -mutant, probably because these specific conditions induced higher rates of H 2 evolution that only the wild type has the capacity of reutilizing through the oxyhydrogen reaction. These findings support the hypothesis that the uptake hydrogenase plays a role in minimizing the loss of energy caused by the nitrogenase-dependent H 2 formation.
Schematic representation of the hupL rearrangement occurring in Nostoc sp. PCC 7120 and other heterocystous cyanobacteria (adapted from Carrasco et al. , 2005 ). In the vegetative cells, hupL is interrupted by a DNA element that is excised late during the heterocyst differentiation process by a site-specific recombination. Subsequently, the structure of the hupL gene is restored, allowing its expression in the heterocysts only. The destiny of the 9.5-kb excised element is currently unknown. In aerobically grown filaments of Nostoc sp. PCC 7120, most of the uptake hydrogenase activity is recovered in the membrane fraction of heterocysts ( Houchins & Burris, 1981b ). The question marks represent events that have not been elucidated so far: the fate of the excised DNA element, and the attachment of the uptake hydrogenase to a cell membrane.
Despite the hupL element being absent from the two other heterocystous strains for which genome sequences are available, A. variabilis and N. punctiforme (see also Oxelfelt et al. , 1998 ; Happe et al. , 2000 ), DNA hybridization studies showed that sequences similar to xisC were present in about half of the heterocystous strains tested ( Tamagnini et al. , 2000 ). These authors also showed that the presence of the bidirectional hydrogenase is not ubiquitous among heterocystous cyanobacteria, although they could not establish a correlation between the presence/absence of the bidirectional enzyme and hupL rearrangement.
hupSL intergenic region
The regions between hupS and hupL in cyanobacteria are longer than in other microorganisms, differ considerably in size (ranging from 43 to 689 bp; see Table 2 ) and are not particularly conserved (except for Nostoc sp. PCC 7120 and A. variabilis ). A prominent feature within the hupSL intergenic region of heterocystous strains is the presence of Short Tandemly Repeated Repetive (STRR) sequences (with the exception of the relatively short 43-bp region of Nostoc sp. Mitsui 38901). STRR sequences have previously been shown to be frequent in heterocyst-forming cyanobacteria and relatively less frequent in unicellular strains ( Asayama et al. , 1996 ). Indeed, no STRR sequences could be discerned in the hupSL intergenic region from nonheterocystous cyanobacteria. However, in the filamentous nonheterocystous L. majuscula only about 10% of the intergenic region consists of nonrepetitive nucleotides, with two distinct sets of Long Repeated Repetitive (LRR) sequences clearly identified (for details see Leitão, 2005 ). Because the repetitive sequences within the hupSL intergenic region are highly variable or even absent ( Table 2 ), it is unlikely that these repeats play a direct role in the regulation of gene expression. However, in all strains, a putative stem-loop structure, derived via 2D-computer modeling, might occur in the transcribed RNA ( Lindberg et al. , 2000 ; Tamagnini et al. , 2002 , 2005 ). The value of free energy (ΔG) was determined for each secondary structure and it was negative in all cases (ranging from −136.32 to −6.9 kcal mol −1 ), meaning that the formation of the hairpin is favored. It has been hypothesized that the occurrence of the hairpin may increase the stability of the transcript, and/or confer a translational coupling between hupS and hupL by sequestering the ribosome-binding site of hupL and thereby preventing the initiation of translation of this gene ( Lindberg et al. , 2000 ). However, although the sequestration of the hupL RBS may be effective in N. punctiforme in which the hairpin folds the entire hupSL intergenic region ( Lindberg et al. , 2000 ), it does not occur in all hupSL intergenic hairpin structures predicted. Only the construction of specific mutants will help to clarify the function of these intergenic regions.
Size and occurrence of repetitive sequences within the region between of hupS and hupL in cyanobacteria
| Organism | Size (bp) | Repetitive sequences | GenBank accession number/Reference |
| Unicellular | |||
| Crocosphaera watsonii WH 8501 | 67 | No | NZ_AADV02000237 |
| Cyanothece sp. ATCC 51142 | 126 | No | DQ650318 |
| Gloeothece sp . ATCC 27152 | 259 | No | AY260103Oliveira, (2004) |
| Filamentous nonheterocystous | |||
| Lyngbya aestuarii CCY 9616 | 118 | No | DQ375444 |
| Lyngbya majuscula CCAP 1446/4 | 643 | LRR | AF368526Leitão, (2005) |
| Trichodesmium erythraeum IMS 101 | 689 | No | NZ_AABK04000005 |
| Filamentous heterocystous | |||
| Anabaena siamensis TISTR8012 | 195 | STRR | AY152844 |
| Anabaena variabilis ATCC 29413 | 75 | STRR | Y13216; NC_007413Happe, (2000) . |
| Nostoc HCC 1048 (Mitsui 38901) | 43 | No | AF455566 |
| Nostoc HCC 1061 (Mitsui 56111) | 118 | STRR | AF455567 |
| Nostoc HCC 1075 (Mitsui 91911) | 97 | STRR | AF455568 |
| Nostoc sp. PCC 7120 | 68 | STRR | U08013 ; NC_003272Carrasco, (1995) , Kaneko, (2001) |
| Nostoc sp. PCC 7422 | 144 | STRR | AB237640 |
| Nostoc muscorum CCAP 1453/12 | 68 | STRR | AF455565 Oxelfelt et al. (1998) |
| Nostoc punctiforme PCC 73102 | 192 | STRR | AF030525; NZ_AAAY02000001Oxelfelt, (1998) |
| Organism | Size (bp) | Repetitive sequences | GenBank accession number/Reference |
| Unicellular | |||
| Crocosphaera watsonii WH 8501 | 67 | No | NZ_AADV02000237 |
| Cyanothece sp. ATCC 51142 | 126 | No | DQ650318 |
| Gloeothece sp . ATCC 27152 | 259 | No | AY260103Oliveira, (2004) |
| Filamentous nonheterocystous | |||
| Lyngbya aestuarii CCY 9616 | 118 | No | DQ375444 |
| Lyngbya majuscula CCAP 1446/4 | 643 | LRR | AF368526Leitão, (2005) |
| Trichodesmium erythraeum IMS 101 | 689 | No | NZ_AABK04000005 |
| Filamentous heterocystous | |||
| Anabaena siamensis TISTR8012 | 195 | STRR | AY152844 |
| Anabaena variabilis ATCC 29413 | 75 | STRR | Y13216; NC_007413Happe, (2000) . |
| Nostoc HCC 1048 (Mitsui 38901) | 43 | No | AF455566 |
| Nostoc HCC 1061 (Mitsui 56111) | 118 | STRR | AF455567 |
| Nostoc HCC 1075 (Mitsui 91911) | 97 | STRR | AF455568 |
| Nostoc sp. PCC 7120 | 68 | STRR | U08013 ; NC_003272Carrasco, (1995) , Kaneko, (2001) |
| Nostoc sp. PCC 7422 | 144 | STRR | AB237640 |
| Nostoc muscorum CCAP 1453/12 | 68 | STRR | AF455565 Oxelfelt et al. (1998) |
| Nostoc punctiforme PCC 73102 | 192 | STRR | AF030525; NZ_AAAY02000001Oxelfelt, (1998) |
LRR, long repeated repetitive; STRR, short tandemly repeated repetitive.
Size and occurrence of repetitive sequences within the region between of hupS and hupL in cyanobacteria
| Organism | Size (bp) | Repetitive sequences | GenBank accession number/Reference |
| Unicellular | |||
| Crocosphaera watsonii WH 8501 | 67 | No | NZ_AADV02000237 |
| Cyanothece sp. ATCC 51142 | 126 | No | DQ650318 |
| Gloeothece sp . ATCC 27152 | 259 | No | AY260103Oliveira, (2004) |
| Filamentous nonheterocystous | |||
| Lyngbya aestuarii CCY 9616 | 118 | No | DQ375444 |
| Lyngbya majuscula CCAP 1446/4 | 643 | LRR | AF368526Leitão, (2005) |
| Trichodesmium erythraeum IMS 101 | 689 | No | NZ_AABK04000005 |
| Filamentous heterocystous | |||
| Anabaena siamensis TISTR8012 | 195 | STRR | AY152844 |
| Anabaena variabilis ATCC 29413 | 75 | STRR | Y13216; NC_007413Happe, (2000) . |
| Nostoc HCC 1048 (Mitsui 38901) | 43 | No | AF455566 |
| Nostoc HCC 1061 (Mitsui 56111) | 118 | STRR | AF455567 |
| Nostoc HCC 1075 (Mitsui 91911) | 97 | STRR | AF455568 |
| Nostoc sp. PCC 7120 | 68 | STRR | U08013 ; NC_003272Carrasco, (1995) , Kaneko, (2001) |
| Nostoc sp. PCC 7422 | 144 | STRR | AB237640 |
| Nostoc muscorum CCAP 1453/12 | 68 | STRR | AF455565 Oxelfelt et al. (1998) |
| Nostoc punctiforme PCC 73102 | 192 | STRR | AF030525; NZ_AAAY02000001Oxelfelt, (1998) |
| Organism | Size (bp) | Repetitive sequences | GenBank accession number/Reference |
| Unicellular | |||
| Crocosphaera watsonii WH 8501 | 67 | No | NZ_AADV02000237 |
| Cyanothece sp. ATCC 51142 | 126 | No | DQ650318 |
| Gloeothece sp . ATCC 27152 | 259 | No | AY260103Oliveira, (2004) |
| Filamentous nonheterocystous | |||
| Lyngbya aestuarii CCY 9616 | 118 | No | DQ375444 |
| Lyngbya majuscula CCAP 1446/4 | 643 | LRR | AF368526Leitão, (2005) |
| Trichodesmium erythraeum IMS 101 | 689 | No | NZ_AABK04000005 |
| Filamentous heterocystous | |||
| Anabaena siamensis TISTR8012 | 195 | STRR | AY152844 |
| Anabaena variabilis ATCC 29413 | 75 | STRR | Y13216; NC_007413Happe, (2000) . |
| Nostoc HCC 1048 (Mitsui 38901) | 43 | No | AF455566 |
| Nostoc HCC 1061 (Mitsui 56111) | 118 | STRR | AF455567 |
| Nostoc HCC 1075 (Mitsui 91911) | 97 | STRR | AF455568 |
| Nostoc sp. PCC 7120 | 68 | STRR | U08013 ; NC_003272Carrasco, (1995) , Kaneko, (2001) |
| Nostoc sp. PCC 7422 | 144 | STRR | AB237640 |
| Nostoc muscorum CCAP 1453/12 | 68 | STRR | AF455565 Oxelfelt et al. (1998) |
| Nostoc punctiforme PCC 73102 | 192 | STRR | AF030525; NZ_AAAY02000001Oxelfelt, (1998) |
LRR, long repeated repetitive; STRR, short tandemly repeated repetitive.
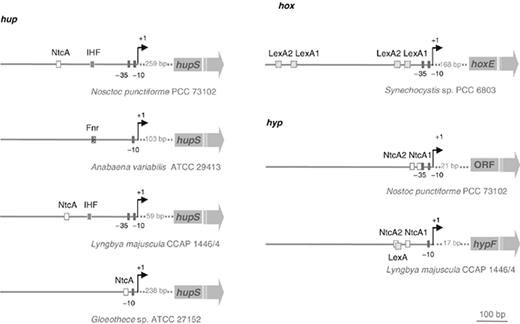
hup promoter regions and transcriptional regulators
As mentioned previously, in all cyanobacteria studied so far the uptake hydrogenase structural genes are arranged in a contiguous manner with the gene encoding the smaller subunit located upstream of the gene of the larger one. The transcriptional start sites of the hup operons are localized 238, 59, 103 and 259 bp upstream from the hupS start codon for the unicellular Gloeothece sp. ATCC 27152, the filamentous L. majuscula and the filamentous heterocystous A. variabilis and N. punctiforme , respectively ( Happe et al. , 2000 ; Lindberg et al. , 2000 ; Oliveira et al. , 2004 ; Leitão, 2005 ) ( Fig. 4 ). The analysis of the regions upstream the transcriptional start point (tsp) revealed the presence of a −10 and a −35 box in both L. majuscula and N. punctiforme , while in Gloeothece sp. ATCC 27152 and A. variabilis only a −10 box could be clearly discerned. A putative binding site for NtcA (a protein that operates global nitrogen control in cyanobacteria) could be found in Gloeothece sp. ATCC 27152, L. majuscula and N. punctiforme , although its relative position to the tsp varied depending on the strain. Moreover, in L. majuscula and N. punctiforme a possible binding site for the integration host factor (IHF) – WATCAAN 4 TTR ( Craig & Nash, 1984 ; Goodrich et al. , 1990 ; Goodman et al. , 1999 ) – could be recognized in the region between the NtcA motif and the tsp ( Fig. 4 ). It has been postulated that the possible binding of the IHF to the promoter could bend the DNA ( Friedman et al. , 1988 ), and consequently allow the contact of the NtcA with the RNA polymerase complex, activating the hupSL transcription. In the unicellular Gloeothece sp. ATCC 27152, the potential NtcA-binding site is centered at −41.5 bp with respect to the tsp in place of the −35 box, like in the canonical NtcA-activated promoters with the consensus sequence signature GTAN 8 TAC ( Herrero et al. , 2001 ), a structure similar to that of class II bacterial promoters activated by catabolite activator protein (CAP). In L. majuscula and N. punctiforme , the NtcA-binding sites were found to be centered at positions −233.5 and −258.5, respectively, resembling class I CAP-dependent promoters ( Busby & Ebright, 1999 ; Herrero et al. , 2001 , 2004 ). These data indicate that the type of the NtcA-activated promoter (class I vs II) is not correlated to the strategies used by heterocystous and nonheterocystous cyanobacteria to separate N 2 fixation and photosynthesis. In the filamentous heterocystous A. variabilis , half of a sequence motif identical to the consensus Fnr-binding sequence was identified 144-bp upstream of the tsp ( Happe et al. , 2000 ) ( Fig. 4 ). Fnr is a regulator of a fumarate nitrate reductase, which has been found to be involved in the regulation of the hyp operon in Escherichia coli ( Lutz et al. , 1991 ), and it is responsible for the induction of several operons in E. coli grown under anaerobic conditions ( Spiro & Guest, 1990 ). In A. variabilis , although there is no rearrangement of the hupL gene, hupSL are expressed in heterocysts only. These differentiated cells have very low intracellular O 2 pressures which led Happe, (2000) to suggest that the hupSL operon in A. variabilis could be regulated in a manner similar to that of the anaerobically induced operons in E. coli .
Promoter regions upstream of hupS , hoxE and hypF in cyanobacteria. The following regions are highlighted: putative NtcA-, IHF-, Fnr- and LexA-binding sites, the −10 and −35 boxes and the transcriptional start points (+1). The following ORFs are not to scale. In Nostoc punctiforme , the ORF represented here is immediately upstream of hypF and in the same direction. Analysis of the available genomes revealed the presence of homologues of this ORF, in the same position and direction, in other filamentous cyanobacteria, and the encoded proteins can be assigned to COG0583 that includes transcriptional regulators from the LysR family ( Leitão, 2006 ). In Synechocystis sp. PCC 6803 hox promoter region, the two putative pairs of LexA-binding motifs were identified by two different groups ( Gutekunst et al. , 2005 ; Oliveira & Lindblad, 2005 ).
The possible interaction between NtcA and the hupSL ( W ) promoter regions in cyanobacteria was assessed by performing band shift assays. These experiments indicate a specific binding of NtcA to DNA sequences upstream of hupS in the three cyanobacterial strains tested ( Gloeothece sp. ATCC 27152, L. majuscula and N. punctiforme ), suggesting, indeed, the involvement of NtcA in the transcription regulation of the uptake hydrogenase gene cluster ( Lindberg et al. , 2003 ; Oliveira et al. , 2004 ; Leitão, 2005 ). The fact that the transcription of the uptake hydrogenase structural genes is under the control of the transcriptional regulator that operates global nitrogen control in cyanobacteria reinforces the correlation observed between the activity of the uptake hydrogenase and N 2 fixation, already demonstrated in several filamentous heterocystous cyanobacteria ( Houchins et al. , 1984 ; Wolk et al. , 1994 ; Oxelfelt et al. , 1995 ; Troshina et al. , 1996 ).
Transcription and expression patterns of hup genes
The first transcriptional data on cyanobacterial uptake hydrogenases arose from RT-PCR experiments on Nostoc sp. PCC 7120, revealing that hupL is expressed only after a photosynthetic vegetative cell differentiates into a N 2 -fixing heterocyst (see above details about the DNA rearrangement occurring within this strain, Carrasco et al. , 1995 , 2005 ). Subsequent studies with other filamentous heterocystous strains have shown that hupSL is a transcriptional unit ( Happe et al. , 2000 ; Lindberg et al. , 2000 ), present in cells grown under N 2 -fixing conditions ( Axelsson et al. , 1999 ; Happe et al. , 2000 ; Hansel et al. , 2001 ). Non-N 2 -fixing cultures of Nostoc muscorum , a strain without the hupL rearrangement, exhibit no in vivo H 2 -uptake activity ( Axelsson et al. , 1999 ). However, the transfer of N. muscorum cells from non-N 2 -fixing (ammonia) to N 2 -fixing conditions induced the appearance of a transcript (after c . 24 h), and the relative amounts of transcript increased in parallel with the H 2 -uptake activity ( Axelsson et al. , 1999 ). A similar pattern of transcription was observed for A. variabilis and N. punctiforme , two other strains with noninterrupted hupL genes ( Happe et al. , 2000 ; Hansel et al. , 2001 ). These authors demonstrated that hupSL transcripts were missing in A. variabilis and in N. punctiforme cells grown with ammonia (and in A. variabilis cells grown with nitrate), but were present in both organisms grown under N 2 -fixing conditions.
While the heterocyst provides a microaerobic environment protecting the oxygen-sensitive nitrogenases and uptake hydrogenases from the atmospheric and intracellulary generated oxygen, the nonheterocystous cyanobacteria developed different approaches. The temporal separation between photosynthesis (light) and nitrogen-fixation/hydrogen uptake (dark) seems to be the most common strategy adopted by the later cyanobacteria ( Bergman et al. , 1997 ; Böhme, 1998 ; Berman-Frank et al. , 2003 ). In fact, in the nonheterocystous Gloeothece sp. ATCC 27152 (unicellular) and L. majuscula (filamentous), grown under nitrogen-fixing conditions and 12 h light/12 h dark cycles, there is an evident light/dark regulation with the highest levels of hupSL ( W ) transcripts detected during the light phase or in the transition between the light and dark phase, respectively ( Oliveira et al. , 2004 ; Leitão, 2005 ). It has also been demonstrated that both organisms exhibit higher hydrogen-uptake activities during the dark period (in agreement with the nitrogen fixation rates; see Reade et al. , 1999 ; Lundgren et al. , 2003 ). In L. majuscula , the increase of the HupL protein levels coincides with the increase of hydrogenase uptake activity during the dark phase. In the beginning of the light phase, no hupSL transcription is detectable, and the levels of both polypeptides and H 2 uptake activity begin to decline ( Leitão, 2005 ). These results suggest that in L. majuscula , a protein turnover occurs, with degradation taking place during the light period and de novo synthesis taking place during the dark phase. The time difference between the hupSL transcription and the hydrogen uptake activity, both in Gloeothece sp. ATCC 27152 and L. majuscula , might be due to the complexity of the maturation process of the uptake hydrogenase. Thus, it is possible that the translation occurs as soon as the transcript is available, while the enzyme becomes active only after the maturation process is completed. The temporal separation between the photosynthesis and nitrogen fixation/hydrogen uptake activity may also influence the time lag between transcription and activity.
In the presence of combined nitrogen, hupSLW transcription is totally repressed in Gloeothece sp. ATCC 27152, while in L. majuscula the levels of hupSL transcription and expression are significantly reduced but it is possible to discern a pattern similar to the one observed in cells grown under N 2 -fixing conditions ( Oliveira et al. , 2004 ; Leitão, 2005 , Ferreira, 2007) . The results obtained for L. majuscula under non-N 2 -fixing conditions could be explained by the mode of growth of this cyanobacterium, in which the inner cells are probably not in the same conditions notably in terms of access to the combined nitrogen.
Besides the source of nitrogen, other factors were proven to influence the transcription/expression of the cyanobacterial uptake hydrogenases. Similar to any NiFe hydrogenase, the activity of the cyanobacterial uptake enzyme was shown to be dependent on nickel availability, and the addition of external nickel to the growth medium (up to a certain concentration) increased the uptake hydrogenase activity in several strains ( Xiankong et al. , 1984 ; Daday et al. , 1985 ; Kumar & Polasa, 1991 ; Oxelfelt et al. , 1995 ; Axelsson & Lindblad, 2002 ). Furthermore, the addition of exogenous hydrogen was shown to induce hupSL transcription and hydrogen uptake activity in N. muscorum and N. punctiforme ( Oxelfelt et al. , 1995 ; Axelsson & Lindblad, 2002 ), as well as hydrogen uptake activity in Nostoc sp. PCC 7120 ( Houchins & Burris, 1981b ). Both cyanobacterial hydrogenases are affected by the oxygen partial pressure. Nostoc muscorum and N. punctiforme cultures transferred from aerobic to anaerobic conditions showed an increase in both the transcription of hupL and hydrogen uptake activity ( Axelsson & Lindblad, 2002 ). Similarly, the uptake hydrogenase activity could be elicited by removing oxygen from the sparging gas of a culture of Nostoc sp. PCC 7120 ( Houchins & Burris, 1981b ). The addition of organic carbon to the culture medium can also influence the hydrogen uptake activity. Cells of N. punctiforme grown either photo- or chemoheterotrophically reach both higher nitrogenase and hydrogen uptake activities than photoautotrophically grown cells ( Oxelfelt et al. , 1995 ). However, the effect of carbon substrates on the cyanobacterial uptake hydrogenase activity is difficult to assess, and apparently contradictory results are reported in the literature ( Houchins, 1984 ; Kumar et al. , 1986 ; Chen et al. , 1989 ; Margheri et al. , 1991 ).
Bidirectional hydrogenase
The soluble or loosely membrane associated cyanobacterial bidirectional hydrogenase might be present in both N 2 - and non-N 2 -fixing strains ( Tamagnini et al. , 2000 , 2002 ). Initially, the bidirectional hydrogenase was thought to be composed of four subunits (encoded by the hox – hydrogen oxidation – genes), in which HoxFU constitute the diaphorase part, and HoxYH constitute the hydrogenase part ( Schmitz et al. , 1995 ; Appel & Schulz, 1996 ; Boison et al. , 1996 , 1998 ; Sheremetieva et al. , 2002 ). However, because HoxE was shown to copurify with the active bidirectional enzyme, the cyanobacterial bidirectional hydrogenase is considered to be a heteropentameric enzyme encoded by hoxEFUYH , HoxE belonging to the diaphorase part ( Schmitz et al. , 2002 ). Bidirectional hydrogenases with more than four subunits have also been identified in other bacteria, such as the photosynthetic purple sulfur bacteria Thiocapsa roseopersicina and Allochromatium vinosum which contain heteropentameric cyanobacterial-type bidirectional hydrogenases ( Rákhely, 2004 ; Long et al. , 2007 ), and Ralstonia eutropha , which possess two HoxI subunits besides HoxFUYH ( Burgdorf et al. , 2005 ). In recent years, the number of reports showing the presence of a functional and active bidirectional hydrogenase in cyanobacteria has increased significantly, ranging from unicellular strains ( Gloeocapsa alpicola CALU 743 – Sheremetieva et al. , 2002 ; Troshina et al. , 2002 ) to filamentous nonheterocystous ( L. majuscula – Schütz, 2004 ; Leitão, 2005 ; Arthrospira and Spirulina spp. – Zhang et al. , 2005a , b ), and filamentous heterocystous strains ( Nostoc spp. – Tamagnini et al. , 2000 ; Schütz, 2004 ). Furthermore, the increasing number of cyanobacterial sequenced genomes is contributing toward a better understanding of both the distribution and the diversity of this enzyme.
The physiological function of the bidirectional hydrogenase in cyanobacteria is not totally clear. It has been suggested that the enzyme acts as an electron valve during photosynthesis in Synechocystis sp. PCC 6803. This is based on the fact that hoxH− mutants are impaired in the oxidation of PSI, have higher fluorescence of PSII and have different transcript levels of the photosynthetic genes psbA , psaA and petB when compared with the wild type ( Appel et al. , 2000 ). The enzyme has also been proposed to play a role in fermentation functioning as a mediator in the release of excess reducing power under anaerobic conditions ( Stal & Moezelaar, 1997 ; Troshina et al. , 2002 ). Furthermore, it has been suggested previously that the bidirectional hydrogenase could be part of the respiratory complex I ( Appel & Schulz, 1996 ; Schmitz & Bothe, 1996 ), because only 11 subunits out of 14 conserved subunits of the prokaryotic complex I have been identified in cyanobacteria. Some of the subunits of the bidirectional hydrogenase indeed show sequence similarities with the missing subunits of the respiratory complex I ( Schmitz et al. , 1995 ). However, the bidirectional hydrogenase has been demonstrated to be absent from several cyanobacterial strains ( Tamagnini et al. , 1997 , 2000 ; Schütz, 2004 ; Ludwig et al. , 2006 ). Moreover, N. punctiforme , a strain naturally lacking the bidirectional hydrogenase ( Tamagnini et al. , 1997 ), has rates of respiration comparable to cyanobacteria containing the bidirectional hydrogenase ( Boison et al. , 1999 ). In addition, mutants of hoxU in Synechococcus sp. PCC 6301 (former Anacystis nidulans ) ( Boison et al. , 1998 ) and hoxEF in Synechocystis sp. PCC 6803 ( Howitt & Vermaas, 1999 ) showed nonimpaired respiratory O 2 uptake while being affected in H 2 evolution. Furthermore, inactivation of hoxH in Synechocystis sp. PCC 6803 and Nostoc sp. PCC 7120 resulted only in a small decrease in the growth rate compared with the respective wild types ( Appel et al. , 2000 ; Masukawa et al. , 2002 ). Taking into account all the data, it seems that in general the bidirectional hydrogenase does not play an essential role for cell survival in the strains where it is present.
Attempting to shed some light on the physiological function of the bidirectional hydrogenase, Cournac, (2004) demonstrated that the bidirectional hydrogenase in Synechocystis sp. PCC 6803 is insensitive to light, reversibly inactivated by O 2 and can be quickly reactivated by NADH or NADPH. This work also reported H 2 evolution by cells incubated anaerobically in the dark, after an adaptation period. This dark H 2 evolution was enhanced by exogenously added glucose and resulted from the oxidation of NAD(P)H produced by fermentation reactions. Upon illumination, a short (<30 s) burst of H 2 output was observed, followed by rapid H 2 uptake, and a concomitant decrease in CO 2 concentration in the cyanobacterial cell suspension, which were both linked to photosynthetic electron transport in the thylakoids ( Cournac et al. , 2004 ). Moreover, in this experimental setup, in anoxia (or microaerobiosis) and in the presence of H 2 , H 2 uptake was of the same magnitude as photosynthetic activity and could therefore contribute significantly to CO 2 fixation. Therefore, although the bidirectional hydrogenase in Synechocystis sp. PCC 6803 is constitutively expressed in the presence of O 2 ( Appel et al. , 2000 ), it probably plays a role mainly under anaerobic or microaerobic conditions, and at the onset of light before the enzyme is inactivated by photosynthetic O 2 . In the ndhB mutant M55, which is defective in the type I NADPH-dehydrogenase complex (NDH-1) and produces only low amounts of O 2 in the light, H 2 uptake was negligible during dark-to-light transitions, allowing several minutes of continuous H 2 production. It was further shown that two pathways of electron supply for H 2 production operate in M55, namely photolysis of water at the level of photosystem II and carbohydrate-mediated reduction of the plastoquinone pool. When comparing the features of the Synechocystis sp. PCC 6803 hydrogenase with those of the homologous NAD + -dependent hydrogenase of R. eutropha , despite sequence homologies between the two enzymes, their characteristics are not identical, which might indicate that this enzyme might have slightly different functions in different organisms ( Cournac et al. , 2004 ).
If the function of the bidirectional hydrogenase is still open to debate, its subcellular localization is not less controversial. The bidirectional hydrogenase can be found in both the heterocysts and the vegetative cells ( Hallenbeck & Benemann, 1978 ; Houchins & Burris, 1981a ), and in Nostoc sp. PCC 7120 appears in the soluble fraction after cell disruption, and consequently has been considered to be a soluble enzyme ( Houchins & Burris, 1981b ). Nevertheless, investigations in other cyanobacteria suggest a weak association of the bidirectional hydrogenase with cell membranes: in A. variabilis and Synechocystis sp. PCC 6803, an association with the thylakoid membrane was proposed ( Serebriakova et al. , 1994 ; Appel et al. , 2000 ), while in Synechococcus sp. PCC 6301 immunological data implied an association with the cytoplasmic membrane ( Kentemich et al. , 1989 , 1991 ).
Physical organization of hox genes and the corresponding proteins
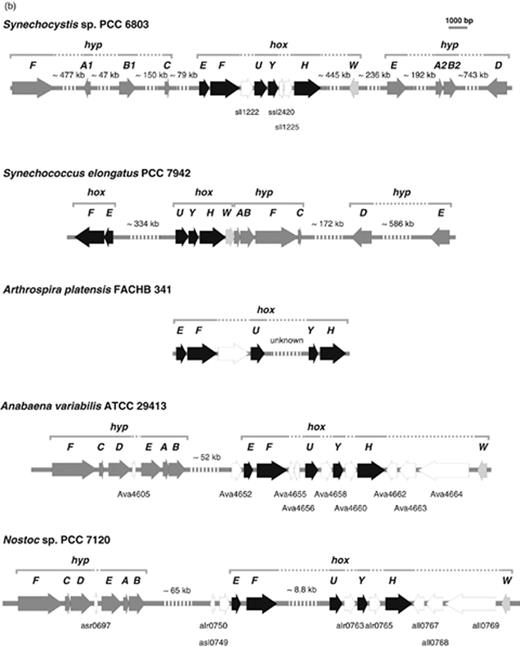
In cyanobacteria, the structural genes encoding the bidirectional hydrogenase are organized in a dissimilar way (see Fig. 2 ). In some strains (e.g. Synechocystis sp. PCC 6803 and A. variabilis ), the hox genes are localized in one cluster, although interspersed with different ORFs at diverse positions. In other cases, the hox genes are found in two different clusters separated by several kilobase ( c . 333 and 8.8 kb in Synechococcus sp. PCC 6301 and Nostoc sp. PCC 7120, respectively). Despite this fact, the similarities at the deduced amino acid level of their homologous hydrogenase proteins range between 55% and 81%.
The bidirectional hydrogenase has been purified from several cyanobacterial strains: A. cylindrica ( Hallenbeck & Benemann, 1978 ), Spirulina maxima ( Llama et al. , 1979 ), Microcystis aeruginosa ( Asada et al. , 1987 ), Synechococcus sp. PCC 6301 ( Schmitz et al. , 1995 , 2002 ) and Synechocystis sp. PCC 6803 ( Schmitz et al. , 2002 ), but the data collected by Schmitz, (2002) finally helped to clarify the picture of the subunit composition and molecular mass of the cyanobacterial bidirectional hydrogenase. Thus, it is widely accepted that the bidirectional hydrogenase is composed of five subunits, HoxE, HoxF, HoxU, HoxY and HoxH, with apparent molecular weights of c . 20, 61, 28, 24 and 49 kDa, respectively. The molecular weight of the native protein (375 kDa) indicates a dimeric assembly of the enzyme complex, Hox(EFUYH) 2 ( Schmitz et al. , 2002 ).
Similar to the uptake hydrogenase, the large subunit of the hydrogenase dimer (HoxH) harbors the active metal center containing nickel and iron. The two metal atoms are held in close proximity by two disulfide bridges provided by two cysteine residues of the protein. The iron has two cyanide ions and one carbon monoxide as ligands, whereas the nickel ion is coordinated by two additional cysteines ( Volbeda et al. , 1995 ). The small subunit of the hydrogenase dimer (HoxY), and the different components of the diaphorase part of the bidirectional hydrogenase (HoxF and HoxU) also contain several conserved cysteine residues putatively involved in the coordination of FeS clusters ( Schmitz et al. , 2002 ; for a review, see Tamagnini et al. , 2002 ). In addition, in the middle region of HoxF, typical glycine-rich binding sites for NAD + (GxGxxGxxxG) and flavin mononucleotide (GxGxxxxGx 10 GxxG) can be identified ( Schmitz et al. , 1995 ). HoxE may be involved as a bridging subunit in membrane attachment. Moreover, a functional role in electron transport directed to membrane components, as demonstrated experimentally for the Hox-hydrogenase of Thiocapsa roseopersicina ( Rákhely, 2004 ), could be considered because sequence motifs for binding of an additional FeS cluster are present in this gene ( Schmitz et al. , 2002 ).
hox promoter regions and transcriptional regulators
The information about the transcription and regulation of the hox genes is limited in cyanobacteria, but the understanding of these mechanisms is now emerging. Recent studies showed that the hox genes in Synechocystis sp. PCC 6803 are transcribed as a single operon ( Gutekunst et al. , 2005 ; Oliveira & Lindblad, 2005 ; Antal et al. , 2006 ) with the transcription start point located 168-bp upstream of the hoxE start codon ( Gutekunst et al. , 2005 ; Oliveira & Lindblad, 2005 ).
Up to now, only one regulator – LexA – has been proven to bind and regulate the transcription of the hox genes in cyanobacteria. Two independent studies ( Gutekunst et al. , 2005 ; Oliveira & Lindblad, 2005 ) demonstrated an interaction between LexA and the promoter region of the bidirectional hydrogenase in Synechocystis sp. PCC 6803. However, two distinct regions were analyzed and both were demonstrated to be targets for this interaction. Oliveira & Lindblad (2005) showed that LexA binds to a region located between the nucleotides −198 and −338 bp, respective to translational start point, while Gutekunst, (2005) found that LexA interacts further upstream on the hox promoter, at the positions −592 to −690 bp, in relation to the hoxE ATG codon (see Fig. 4 ). Furthermore, a LexA-depleted mutant showed a reduced hydrogenase activity compared with the wild-type, suggesting that LexA works as a transcription activator of the hox genes in Synechocystis sp. PCC 6803 ( Gutekunst et al. , 2005 ). Synechocystis sp. PCC 6803 LexA has been detected in different proteomic studies ( Wang et al. , 2000 ; Gan et al. , 2005 ; Srivastava et al. , 2005 ; Fulda et al. , 2006 ; Kurian et al. , 2006 ; Slabas et al. , 2006 ), and its transcript has also been identified in microarray experiments ( Hihara et al. , 2001 ; Kamei et al. , 2001 ; Li et al. , 2004 ; Singh et al. , 2004 ; Tu et al. , 2004 ; Shapiguzov et al. , 2005 ). Interestingly, in some proteomic studies, LexA has been identified in association with thylakoid membrane fractions ( Wang et al. , 2000 ; Srivastava et al. , 2005 ), which represents an unexpected location for a transcription regulator.
Based on the observations that the bidirectional hydrogenase activity is directly affected by the redox status of the cell, either in photosynthesis or in fermentation, and that the regulation of the hox gene expression can be operated by LexA, hypothesis was recently put forward on the direct involvement of the transcription regulator LexA as a mediator of the redox-responsive regulation of the hox gene expression in Synechocystis sp. PCC 6803 ( Antal et al. , 2006 ). Interestingly, the expression of the cyanobacterial DEAD-box RNA helicase, crhR , which is regulated in response to conditions that elicit reduction of the photosynthetic electron transport chain, was recently shown as being directly controlled by LexA in Synechocystis sp. PCC 6803 ( Patterson-Fortin et al. , 2006 ). Transcript analysis indicated that lexA and crhR are divergently expressed, with the respective transcripts accumulating differently under conditions, which, respectively, oxidize and reduce the electron transport chain, suggesting that LexA works as a repressor of the crhR transcription ( Patterson-Fortin et al. , 2006 ). Although these results are in agreement with the initial hypothesis, the signal transduction pathways directly or indirectly involved in the regulation of LexA, and consequently its downstream targets, definitely require further investigation.
Transcription and expression patterns of hox genes
The number of studies focusing on the transcription and regulation of the hox genes in cyanobacteria is scarce. Nevertheless, transcripts of the bidirectional hydrogenase have been shown to be present in NH 4+ -grown filaments, and in both vegetative cells and heterocysts under nitrogen-fixing conditions in A. variabilis ( Boison et al. , 2000 ). In addition, hoxFUYH were shown to be transcribed as a single unit together with other two ORFs with unknown function. However, it should be kept in mind that these experiments were performed using RT-PCR and do not exclude additional promoters within the operon ( Boison et al. , 2000 ). On the other hand, in the unicellular Synechococcus sp. PCC 6301 and Synechococcus sp. PCC 7942 the hox genes are located apart and give rise to two different transcripts ( Boison et al. , 2000 ; Schmitz et al. , 2001 ). While hoxEF are cotranscribed in both strains, the second transcript is constituted by hoxUYH together with hoxW , hypA and hypB in Synechococcus sp. PCC 6301 ( Boison et al. , 2000 ), and by hoxUYHW only in Synechococcus sp. PCC 7942 ( Schmitz et al. , 2001 ). For the last strain, using real-time PCR and reporter gene constructs, it was suggested that a second promoter might be present between hoxH and hoxW ( Schmitz et al. , 2001 ). Furthermore, it was demonstrated that the hox genes have a circadian clock expression ( Schmitz et al. , 2001 ), a fact that has also been demonstrated for hoxE in Synechocystis sp. PCC 6803 ( Kucho et al. , 2005 ).
Very few studies focusing on the regulation of hox genes transcription have been performed in cyanobacteria. Analysis of the transcription of hoxY and hoxH in G. alpicola , under combined nitrogen-limiting growth conditions, demonstrated an increase in the enzyme activity, but no regulation at the transcript level ( Sheremetieva et al. , 2002 ). In contrast, Northern blot analyses of the hox genes expression in Synechocystis sp. PCC 6803 under combined nitrogen-limiting growth conditions demonstrated an increase in transcription ( Antal et al. , 2006 ), followed by an increase in enzyme activity (T.K. Antal, P. Oliveira & P. Lindblad, unpublished data). A similar increase in the hox genes transcription has also been observed with microarray in Synechocystis sp. PCC 6803 cells undergoing nitrogen starvation for 4 h ( Osanai et al. , 2006 – supplementary material).
Furthermore, a transfer to a low level of oxygen in A. variabilis induced both the enzyme activity as well as the relative amount of hoxH ( Sheremetieva et al. , 2002 ). It has long been demonstrated that microaerobic/anaerobic conditions influence hox transcription and bidirectional hydrogenase activity in heterocystous cyanobacteria ( Houchins & Burris, 1981a ; Houchins et al. , 1984 ; Serebryakova et al. , 1994 ; Schmitz & Bothe, 1996 ; Axelsson & Lindblad, 2002 ; Sheremetieva et al. , 2002 ). The bidirectional hydrogenase in Nostoc sp. PCC 7120 is active in both vegetative cells and in heterocysts in aerobically grown filaments, with heterocysts having several fold more activity than vegetative cells. When the filaments were transferred to anaerobic conditions, the activity of the bidirectional hydrogenase increased by about two orders of magnitude with approximately the same activity levels in both types of cells ( Houchins & Burris, 1981a ). Similar results have been observed in A. variabilis ( Serebryakova et al. , 1994 ). In contrast to the filamentous cyanobacteria, the activity of the bidirectional hydrogenase in the unicellular G. alpicola is not directly dependent on oxygen ( Troshina et al. , 2002 ). Higher activity is observed under nitrogen starvation and low light, and it was suggested that the bidirectional hydrogenase could act as an alternative electron donor to PSI after inactivation of PSII due to nitrogen starvation. Under dark anoxic conditions, the unicellular cyanobacterium G. alpicola produces H 2 catalyzed by the bidirectional hydrogenase ( Troshina et al. , 2002 ). In addition, the unicellular strain Chroococcidiopsis thermalis CALU 758 contains a bidirectional hydrogenase with some catalytic properties more related to an uptake hydrogenase, i.e. not inducible under anaerobic conditions or under nitrate-starving conditions ( Serebryakova et al. , 2000 ).
Because the bidirectional hydrogenase in cyanobacteria is a metal-dependent enzyme, containing nickel and iron in its active center and FeS clusters involved in electron transfer, the availability of these elements in the growing medium has been a subject of research. Axelsson & Lindblad (2002) showed that in the heterocystous N. muscorum CCAP 1453/12, the addition of external nickel to the growing medium increased the mRNA abundance of hoxH (monitored by RT-PCR). Making use of reporter gene constructs, Gutekunst, (2006) were able to show that the transcription of the bidirectional hydrogenase genes in Synechocystis sp. PCC 6803 increased with lower concentrations of iron, the signal being 10 times higher in cells grown with 0.22 μM iron compared with nonstarved cells. In the same work, measurements of the hydrogenase activity revealed a reduction of the enzyme activity alongside the decrease in the iron concentration. The increase in transcription of the hox genes, when the cells undergo iron starvation, might be a feedback mechanism to compensate for the lack of functionally active enzyme ( Gutekunst et al. , 2006 ). The availability of sulfur in the growth medium has also been shown to influence the bidirectional hydrogenase activity in Synechocystis sp. PCC 6803 and G. alpicola ( Antal & Lindblad, 2005 ). Both strains showed an enhanced (more than fourfold) H 2 production capacity during fermentation via hydrogenase, when grown under sulfur starvation conditions.
Although the understanding of the regulation and the physiological role of the bidirectional hydrogenase is becoming clearer, intriguing recent results on the hydrogenase activity from two substrains of Synechocystis sp. PCC 6803 have shown that they do not have comparable values ( Gutekunst et al. , 2006 ). The authors suggested that these phenotypic differences in the hydrogenase activity might be due to divergences in their metabolism. In fact, maintenance of these strains in culture collections, or under various laboratory conditions, may have led to spontaneous mutations and unintended selective pressures, resulting in the observable variations in each subculture ( Ikeuchi & Tabata, 2001 ). Therefore, special care must be taken when interpreting results coming from different laboratories and different cyanobacterial strains, even from the same strain, but cultured in different laboratories.
Maturation of cyanobacterial hydrogenases
The biosynthesis/maturation of NiFe-hydrogenases is a highly complex process requiring at least seven core proteins for the incorporation of the metal ions and CO and CN ligands in to the active center, the orientation of the FeS clusters within the small subunit and the cleavage of the C-terminus as the final step in the maturation of the large subunit (for a recent review on this subject, see Böck, 2006 , and also Casalot & Rousset, 2001 ; Blokesch et al. , 2002 ; Mulrooney & Hausinger, 2003 ; Kuchar & Hausinger 2004 ; Vignais & Colbeau, 2004 ; Theodoratou et al. , 2005 ). The genes encoding the proteins involved in the maturation of hydrogenases were firstly characterized for E. coli , and while most of the Hyp proteins affect hydrogenases pleiotropically, the large subunit of each hydrogenase is proteolytically processed by a specific endopeptidase ( Lutz et al. , 1991 ; Jacobi et al. , 1992 ; Menon et al. , 1994 ; Rossmann et al. , 1995 ; Theodoratou et al. , 2005 ; Böck, 2006 ). Homologues of the hyp genes are present in all organisms capable of forming NiFe hydrogenases. Although little is known about the biosynthesis/maturation of the cyanobacterial hydrogenases, several genes presumably involved in this process have been identified clustered or scattered throughout the genomes of several cyanobacterial strains ( Boison et al. , 1996 ; Gubili & Borthakur, 1996 , 1998 ; Kaneko et al. , 1996 ; Sakamoto et al. , 1998 ; Hansel et al. , 2001 ; Tamagnini et al. , 2002 ; Wünschiers, 2003 ; Hoffmann et al. , 2006 ; Leitão, 2006 ). The presence of a single copy of most of the hyp genes ( hypFCDEAB ) in the genome of cyanobacteria, regardless of possessing only the uptake hydrogenase (e.g. N. punctiforme ), the bidirectional hydrogenase (e.g. Synechocystis sp. PCC 6803) or both enzymes (e.g. Nostoc sp. PCC 7120) suggests that they might be responsible for the maturation of both hydrogenases. In contrast, the genes encoding for the putative hydrogenase C-terminal endopeptidases – hupW and hoxW – were identified and seem to be specific for the cyanobacterial uptake and the bidirectional hydrogenase, respectively, resembling the situation in other organisms ( Wünschiers, 2003 ; Oliveira et al. , 2004 ; Leitão, 2006 ).
Physical organization of hyp genes and the corresponding proteins
The hyp genes in cyanobacteria are frequently clustered and in the vicinity of the structural genes of one of the hydrogenases ( Fig. 2 ), with a well-known exception – the unicellular non-N 2 -fixing Synechocystis sp. PCC 6803 – in which the hypABCDEF genes are scattered throughout the genome. Still, in this organism the homologs hypA2 and hypB2 are clustered ( Kaneko et al. , 1996 ), but these two do not seem to play a key role in the maturation of the bidirectional hydrogenase ( Hoffmann et al. , 2006 ). In three Synechococcus , closely related strains ( Synechococcus elongatus PCC 6301, Synechococcus elongatus PCC 7942 and Synechococcus sp. PCC 7002) hypABFC are together and downstream of hox genes, while hypD and hypE are apart in the two first organisms ( Boison et al. , 1996 ). In the heterocystous strains, N. punctiforme , Nostoc sp. PCC 7120 and A. variabilis and in the N 2 -fixing but nonheterocystous L. majuscula , the hyp genes are located in a cluster with all genes orientd in the same direction, and relatively close to the uptake hydrogenase structural genes, although in the opposite direction ( Gubili & Borthakur, 1998 ; Hansel et al. , 2001 ; Leitão, 2006 ). However, this organization does not constitute a pattern for N 2 -fixing strains, because it contrasts with the organization observed for other nonheterocystous strains, such as the filamentous T. erythraeum , in which hyp genes are located much further upstream of hupSL (ca. 589 kb), and the unicellular Crocosphaera watsonii WH 8501, in which the genes are scattered over the genome resembling the non-N 2 -fixing Synechocystis sp. PCC 6803. When the genes are grouped, the order varies in non-N 2 -fixing compared with N 2 -fixing strains being hypABFC and hypFCDEAB , respectively. In the former case, ORFs interspersed with the hyp genes can be found in several organisms.
The putative cyanobacterial Hyp proteins possess conserved motifs and may fulfill functions similar to the corresponding proteins in other organisms ( Tamagnini et al. , 2002 ; Vignais & Colbeau, 2004 ; Hoffmann et al. , 2006 ; Leitão, 2006 ). It is believed that from the two metal ions present in the active center of a NiFe hydrogenase, Fe is the first to be incorporated into the enzyme. HypF and HypE are the proteins involved in the synthesis of the CN, and maybe the CO, ligands of iron ( Paschos et al. , 2001 , 2002 ; Böck, 2006 ). HypF accepts carbamoyl phosphate (CP) as a substrate, catalyzes a CP-dependent hydrolysis of ATP into AMP and inorganic phosphate (PPi) and forms an adenylated CP derivative. The carbamoyl group of CP is transferred to the cysteine at the C-terminus of HypE ( Paschos et al. , 2002 ; Reissmann et al. , 2003 ). It was demonstrated in vitro that the CN group from HypE-thiocyanate can be transferred to the complex HypC plus HypD ( Blokesch et al. , 2004a ). Because the transfer of the ligands to the iron requires the input of two electrons ( Blokesch & Bock, 2002 ), HypD is proposed to be the one involved in this process, given that among all the maturation proteins it is the only one with a redox-active cofactor ( Blokesch et al. , 2004a ; Roseboom et al. , 2005 ). On the other hand, HypC is a small chaperone-like protein that was shown to form a complex with HypD ( Blokesch & Bock, 2002 ) and to interact with the large subunit of the hydrogenase ( Magalon & Bock 2000a ; Casalot & Rousset 2001 ). Probably, the liganding of the iron takes place at the HypC–HypD complex ( Blokesch et al. , 2002 , 2004a ), and the interaction between HypC and the precursor of the large subunit leads to the liberation of HypD ( Blokesch et al. , 2004a ; Blokesch & Böck, 2006 ). Subsequently, the liganded Fe is transferred to the precursor of the hydrogenase large subunit ( Blokesch & Böck, 2006 ), and HypC remains attached to the large subunit, maintaining it in an open conformation, allowing the insertion of nickel. This step requires the presence of HypA and HypB ( Jacobi et al. , 1992 ; Olson et al. , 2001 ). HypA is a zinc-containing protein that binds nickel ( Mehta et al. , 2003 ; Blokesch et al. , 2004b ), and HypB is a GTPase that probably plays a dual function: nickel storage and nickel insertation ( Maier, 1993, 1995 ). It is thought that HypA functions as a nickel chaperone and that HypB acts as a regulator, controlling the donation of the metal to the apoprotein or the release of the nickel-free chaperone ( Blokesch et al. , 2004b ). After both metals have been coordinated to the precursor of the large subunit, the C–terminal extension is accessible and can be removed by the specific endopeptidase. The cleavage can only occur after HypC dissociation from the precursor of the large subunit that already contains Ni and Fe(CO)(CN − ) 2 centers ( Magalon & Bock, 2000a, b ), because the endopeptidase uses Ni as a recognition motif. Following the cleavage of the C-terminal tail from the large hydrogenase subunit, the mature large subunit can be assembled, with the mature small subunit forming the functional enzyme ( Magalon & Bock, 2000a ). Maturation of the small subunit should occur in parallel, and independently from the large subunit maturation. The knowledge about this process is still scarce, although recent studies highlighted at least four gene products (encoded within the hup cluster, and downstream of uptake hydrogenase structural genes) that are required for the maturation of the small subunit of the NiFe hydrogenases of Rhizobium leguminosarum bv. viciae ( Manyani et al. , 2005 ; Böck, 2006 ). In cyanobacteria, several additional ORFs are commonly present near hyp or hup genes ( Leitão, 2006 ). The consistent location of these ORFs might indicate that their proteins may have a role in the uptake hydrogenase maturation process and/or its regulation, notably regarding the small subunit.
hyp promoter regions and transcriptional regulators
As mentioned above, the hyp genes can be found clustered or scattered throughout the genome of cyanobacteria ( Fig. 2 ). Analysis of the hyp cluster promoter region of N. punctiforme revealed the presence of −10 and −35 elements, and putative binding sites for NtcA ( Hansel et al. , 2001 ; Fig. 4 ). Similarly, in the corresponding region of L. majuscula the presence of a −10 box, and two putative NtcA-binding sites could be identified. In this organism, a clear −35 box is not present, but it should be taken into account that its sequence is highly variable. Furthermore, a putative LexA-binding site was also found in L. majuscula ( Leitão, 2006 ; Fig. 4 ). The transcriptional regulators NtcA and LexA were shown to bind to the promoter regions of the hup and the hox genes, suggesting their involvement in the regulation of the uptake and bidirectional hydrogenase, respectively (see above, Lindberg et al. , 2003 ; Oliveira et al. , 2004 ; Gutekunst et al. , 2005 ; Leitão, 2005 ; Oliveira & Lindblad, 2005 ). The presence of putative binding sites for both transcriptional factors NtcA and LexA within the hyp operon promoter region, and preliminary results from electrophoretic mobility shift assays ( Ferreira et al. , 2007 ) suggest the involvement of these proteins in the transcriptional regulation of hyp genes in L. majuscula , a cyanobacterium containing both hydrogenases. These data reinforce the hypothesis that the Hyp proteins might be implicated in the maturation/regulation of both hydrogenases, and raise the hypothesis that the transcription of hyp genes in cyanobacteria containing both hydrogenases could be under the control of different transcriptional regulators, e.g. NtcA and LexA.
Transcription and expression patterns of hyp genes
In the heterocystous N. punctiforme , the hup and hyp genes are transcribed under N 2 -fixing but not under non-N 2 -fixing growth conditions ( Hansel et al. , 2001 ). One should bear in mind that N. punctiforme contains only one hydrogenase (the uptake enzyme), and that in this organism both the transcription of hupL and the H 2 uptake activity are repressed when combined nitrogen is present in the growth medium ( Oxelfelt et al. , 1995 ; Hansel et al. , 2001 ).
In the unicellular non-N 2 -fixing Synechocystis sp. PCC 6803, a cyanobacterium harboring only the bidirectional hydrogenase, deletion and insertion mutants of hypA1 , B1 , C , D , E and F showed no hydrogenase activity. Moreover, the complementation of each of the above hyp - inactivated genes restored the bidirectional hydrogenase activity to the wild-type level in the respective mutants ( Hoffmann et al. , 2006 ). In contrast, the deletion of the homologues hypA2 and hypB2 had no effect on the bidirectional hydrogenase activity even though they are transcribed in the wild type, demonstrating that the products of these genes are not actively involved in the maturation process of the bidirectional hydrogenase ( Hoffmann et al. , 2006 ).
Hydrogenase-specific endopeptidases genes hupW and hoxW , and corresponding proteins
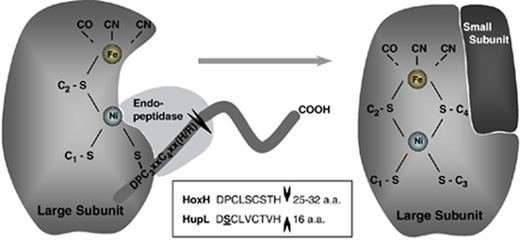
The last step in the processing of the large subunit of NiFe-hydrogenases is the cleavage of a C-terminal peptide, which, most likely, allows a structural reorganization of the molecule and the consequent assembly of the holoenzyme. After both metals have been inserted into the apoprotein precursor of the large subunit, the C-terminal extension is accessible and can be removed by the specific endopeptidase ( Theodoratou et al. , 2005 ; Böck, 2006 ). This process triggers a conformational switch in which the free thiol of the most C-terminally located cysteine residue closes the bridge between the two metals resulting in the formation of the complete heterobinuclear center ( Maier & Bock, 1996 ; Magalon & Bock, 2000a ; Theodoratou et al. , 2005 ; Böck, 2006 ). The peptidase cleaves the hydrogenase large subunit precursor after a histidine or an arginine residue at the C-terminal consensus motif DPCxxCxx(H/R), liberating a short polypeptide that varies considerably both in length and sequence among different organisms ( Wünschiers, 2003 ). It has been postulated that the endopeptidase recognizes its substrate, the nickel-containing hydrogenase precursor, at least in part via the metal that is coordinated by three thiolates, and binds to the exposed C-terminal domain ( Theodoratou et al. , 2000a , b, 2005 and Fig. 5 ). In addition, the endopeptidase interacts with a structural domain to which both the mature part of the large subunit and the C-terminal extension contribute. Therefore, it is believed that the recognition of the hydrogenase by the endopeptidase does not depend on the cleavage site consensus sequence but is mediated by the overall three-dimensional hydrogenase and peptidase protein structures ( Theodoratou et al. , 2000a , b ). After the proteolytic cleavage, the mature large hydrogenase subunit assembles with the small subunit and eventually the enzyme becomes active.
Schematic representation of the putative final step of the maturation process of the Ni–Fe hydrogenases large subunit: cleavage of a small peptide by a specific endopeptidase, followed by a conformational change that encloses the bimetallic center. This structural reorganization of the large subunit will allow the consequent assembly of the holoenzyme. In the large subunits of cyanobacterial hydrogenases – HoxH (bidirectional hydrogenase) and HupL (uptake hydrogenase) – the C-terminal consensus motif DPCxxCxx(H/R) was found in all the deduced sequences, but in HupL the proline is exchanged by a serine (see box). The putative cleaved polypeptide varies in length and sequence for HoxH, while for HupL is always has the same length and is highly conserved.
In cyanobacteria hydrogenase large subunits, the C-terminal consensus motif [DPCxxCxx(H/R)] was found in all the deduced amino acid sequences; however, in the uptake hydrogenase large subunits (HupL) the neutral proline (P) at position 2 of the cutting site motif is exchanged for an uncharged polar serine (S). The sequence of the cutting site motif is totally conserved for each of the cyanobacterial hydrogenases large subunits: HoxH (bidirectional hydrogenase) – DPCLSCSTH; HupL (uptake hydrogenase) – DSCLVCTVH (see Wünschiers, 2003 and Fig. 5 ). The putative cleaved C-terminal polypeptide varies in length (25–32 aa residues) and sequence (10–96% similarity) for HoxH, while for HupL the polypeptide always has the same length (16 aa residues) and is highly conserved for all the deduced sequences [AHDAKTG(E/K)ELARFRT(A/N/S)].
In cyanobacteria, the genes encoding for the putative hydrogenase-specific C-terminal endopeptidases were identified and named hupW and hoxW for the gene encoding the enzyme processing the uptake and the bidirectional hydrogenase, respectively ( Kaneko et al. , 1995 , 2001 ; Boison et al. , 2000 ; Schmitz et al. , 2001 ; Wünschiers, 2003 ; Oliveira et al. , 2004 ; Leitão, 2005 ).
The position of hupW and hoxW in the cyanobacterial chromosome is rather variable; however, in several cases hupW is in the vicinity and in the same direction of hupSL (uptake hydrogenase structural genes). In the nonheterocystous Gloeothece sp. ATCC 27152 and T. erythraeum , hupW is even the ORF located immediately downstream of hupL , and was shown to be cotranscribed with hupSL in Gloeothece sp. ATCC 27152 ( Oliveira et al. , 2004 ). In contrast, in the heterocystous strains A. variabilis , Nostoc sp. PCC 7120 and N. punctiforme , hupW is not part of any known hydrogenase cluster ( Fig. 2 ), and it was shown to be transcribed under N 2 - and non-N 2 -fixing conditions in the last two strains ( Wünschiers, 2003 ). These authors postulated that the transcription of hupW under conditions in which the transcripts of the uptake hydrogenase structural genes could not be detected (presence of ammonia) could imply that hupW is constitutively expressed. Taking into account all the available data, it is not yet possible to establish whether the expression of hupW is or is not constitutive or whether this depends on the strain/existence of cell differentiation.
Similar to what happens for hupW , analysis of the available cyanobacterial genomic sequences revealed that the position and orientation of hoxW in the chromosome is also variable but, in most of the cases, hoxW is downstream of hoxH , one of the bidirectional hydrogenase structural genes ( Fig. 2 ). RT-PCR experiments indicate that in the unicellular non-N 2 -fixing Synechococcus sp. PCC 6301, hoxW is part of a polycistronic message containing hoxUYHWhypAB ( Boison et al. , 2000 ), while in Synechococcus sp. PCC 7942 it was demonstrated that although hoxW constitute a unit together with hoxUYH , it is mainly expressed by its own promoter ( Schmitz et al. , 2001 ). In the heterocystous Nostoc sp. PCC 7120, similar to hupW , hoxW is transcribed under both N 2 - and non-N 2 -fixing conditions ( Wünschiers, 2003 ). Although some data indicate that endopeptidases transcripts are present when the corresponding hydrogenase large subunit transcript is absent, and it has been proposed that their expression is independently regulated from the expression of both the hydrogenase structural and the other accessory genes in cyanobacteria ( Wünschiers, 2003 ), it is premature to make any general conclusion.
To date, two different hydrogenase specific-endopeptidases have been purified and studied, namely HycI and HybD from E. coli ( Rossmann et al. , 1995 ; Fritsche et al. , 1999 ). Both are monomeric proteins of a molecular mass of c . 17 kDa, and they are devoid of metal or other cofactors. Alignments of the amino acid sequences showed that hydrogenase- specific C-terminal endopeptidases share low sequence similarity, with only a few positions fully conserved ( Theodoratou et al. , 2005 ). As a general feature, they have three highly conserved amino acid residues (Glu, Asp and His) that, most likely, have the function of interacting with the nickel in the hydrogenase large subunit precursor ( Theodoratou et al. , 2000a ). The alignment of the putative cyanobacterial endopeptidases with the corresponding proteins from E. coli clearly shows that although the amino acid sequence identity is low, they are indeed structurally related (67–77% structural identity) ( Wünschiers, 2003 ).
Phylogenetic analysis
Recently, the phylogenetic origin of cyanobacterial and algal hydrogenases was analyzed ( Ludwig et al. , 2006 ), leading to the conclusion that Chloroflexus is probably the closest ancestor of cyanobacteria. In all cyanobacterial genomes sequenced to date, and in the genome of Chloroflexus , the two hydrogenase operons – hup and hox – are widely separated on the chromosome, rendering simultaneous gene transfer unlikely. The authors claim that the current distribution of the hydrogenases in cyanobacterial strains probably reflects a differential loss of the genes from their last common ancestor, and that the two sets of genes, encoding the uptake and bidirectional, were either kept in the genome or lost differentially in the different strains according to their ecological needs or constraints. Although the phylogenetic analysis of Ludwig, (2006) clearly demonstrated the monophyly of cyanobacteria, and their relationship with other photosynthetic bacteria, relationships within the cyanobacteria were poorly resolved using HupL sequences. This is probably related to the difficulties of aligning the cyanobacteria with the other highly divergent lineages, and acerbated by the low number of sequences available and long branches leading to terminal nodes. The high variability of the sequences also means that more distant bacterial outgroups cannot be unambiguously aligned. However, analysis solely within the cyanobacteria for both HupS and HupL is less complex, because analysis of the predicted proteins demonstrates that in HupS the number of residues in all known cyanobacteria is constant (320 aa), while HupL generally has 531 aa, with the exception of the filamentous nonheterocystous strains L. majuscula and L. aestuarii with six extra (one insertion of 5 aa and another of one), and T. erythraeum with three extra, coinciding with the position of the five inserts in Lyngbya spp. Owing to this relatively conserved identity, alignment of the amino acids for phylogenetic analysis was facile.
Amino acid sequences were analyzed under the criterion of maximum parsimony, with gaps treated separately as either missing data or as a fifth state. Support for nodes was estimated by bootstrapping with 10 000 replicates.
Both analyses gave widely congruent estimates of phylogeny ( Fig. 6 ). The three heterocystous strains form a clade with 100% support, separated from the nonheterocystous strains by between 16% and 21% divergence. Within the nonheterocystous strains, two pairs of taxa – Cyanothece with Crocosphaera and the two Lyngbya species are well supported. Other relationships are poorly supported. Although the analysis with gaps treated as missing data suggests that the filamentous taxa are not a clade, analysis with gaps treated as a fifth character supported a relationship between T. erythraeum and Lyngbya spp., although with weak support (51%). Thus, exact relationships within this group cannot be ascertained by these sequences, although a sister-taxa relationship between T. erythraeum and L. majuscula is strongly supported through analysis of the Hyp sequences. These results are not in conflict with those suggested by Ludwig (2006) , in which the only well-supported node within cyanobacteria is that of the three heterocystous strains. Evidence for the position of the ancestral root within the cyanobacteria is weak, although T. erythraeum may be sister taxa to the remaining sampled cyanobacteria ( Ludwig et al. , 2006 ).
Unrooted single most parsimonious tree recovered from an MP analysis with gaps treated as missing data of combined small (HupS) and large (HupL) subunit amino acid sequences of cyanobacterial uptake hydrogenases. 210 characters were parsimony informative, and a single tree of 552 steps was recovered (CI=0.83, RI=0.76). NJ recovered an identical topology. Treating gaps as a fifth state altered the topology as indicated in the text. Values beside nodes indicate bootstrap support for MP/NJ. 100 * indicates 100% support in both analyses.
Phylogenetic analysis of cyanobacterial hydrogenases accessory proteins (Hyp A,B,C,D,E and F) and the bidirectional hydrogenases structural proteins (Hox) is complicated by the higher level of variation between species, and in particular greater length variation that leads to uncertain alignment for many positions. Further, not all of the amino acid sequences of hydrogenase accessory proteins are available for all the species analyzed for the uptake hydrogenase structural genes. However, unweighted parsimony analyses indicate that supported estimates of relationships recovered for these proteins do not conflict with the estimate of phylogeny shown in Fig. 6 (analyses not shown).
Genetic engineering/cyanobacterial H 2 production
Cyanobacteria can be used for the production of molecular hydrogen (H 2 ), a possible future energy carrier, which has been the subject of several recent reviews ( Levin et al. , 2004 ; Dutta et al. , 2005 ; Kruse et al. , 2005 ; Prince & Kheshgi, 2005 ; Sakurai & Masukawa, 2007 ). As the main advantages, cyanobacteria can use sunlight as an energy source, water as an electron source and air as a carbon (CO 2 ) and a nitrogen (N 2 ) source. Therefore, no complicated or expensive media are needed for the cultivation of cyanobacteria, and the overall theoretical energy conversion efficiency (from solar energy sun to H 2 ) may be the highest possible.
In cyanobacteria, two natural pathways for H 2 production can be used.
H 2 production as a by-product during nitrogen fixation by nitrogenases
In N 2 -fixing strains, H 2 is produced as a by-product by the nitrogenase enzymatic complex. As this reaction needs the input of ATP (at least two ATP per electron), the overall energy efficiency for hydrogen production is rather low. The turnover of the nitrogenase enzyme is not very high (<10 s −1 ), and the H 2 produced is efficiently taken up by an uptake hydrogenase. The overall oxygen sensitive N 2 -fixation process is occurring in an anaerobic environment achieved using a number of different strategies including spatial or/and temporal separation of N 2 fixation and oxygenic photosynthesis and increased respiration.
It has been reported that n is 1 for the molybdenum-containing enzymes, 3 for the vanadium-nitrogenases and 7.5 for the iron-only nitrogenases, respectively. As a consequence, the alternative nitrogenases, although still very little is known in cyanobacteria, may be better H 2 producers compared with the more conventional molybdenum-nitrogenases.
H 2 production by the bidirectional hydrogenase
The cyanobacterial bidirectional hydrogenase may, under anaerobic conditions, produce and evolve significant amounts of H 2 . Because this reaction is not dependent on ATP, it is energetically more efficient and favorable for H 2 production, with a much higher turnover (1 million turnovers per second) compared with the nitrogenase-based H 2 production. At the same time, the enzyme is not specifically located in an oxygen-protected environment, and the reaction turns into the opposite direction (H 2 uptake) above a certain H 2 partial pressure. Therefore, a continuous and very effective removal of both O 2 and H 2 from the cells and the culture is necessary to lower the overall energy conversion efficiency significantly. Furthermore, an accumulation of ATP could inhibit the electron flow, because it is produced during the linear or cyclic electron flow around PSI, but is not used by the electron acceptor hydrogenase.
Besides the specific challenges for H 2 production connected to the H 2 -evolving enzymes, there are additional unsolved issues for photoautotrophical H 2 production in general. These are related to the low quantum efficiency, due to the naturally large antenna systems of the photosystems, and to electron consuming pathways directly competing with e.g. nitrogenases and hydrogenases.
In summary, to achieve a sustainable, renewable cyanobacterial-based H 2 production, the following challenges have to be addressed:
- 1
efficient H 2 uptake by the cells,
- 2
low energy efficiency and turnover of the nitrogenase and/or the hydrogenase,
- 3
limiting amounts of active H 2 -evolving enzymes,
- 4
high oxygen sensitivity of the nitrogenase and/or the hydrogenase,
- 5
electron flow inhibition by accumulation of ATP in a hydrogenase-driven system,
- 6
low quantum efficiency due to too large antennas in both Photosystem II (PSII) and PSI and
- 7
electron-consuming pathways competing with an efficient electron transfer to the H 2 enzymes.
In recent years, there have been attempts to overcome these barriers and problems, mainly by targeted genetic engineering of cyanobacterial strains:
(1) Efficient H2uptake by the cells : Cyanobacteria have evolved an effective mechanism to recycle the H 2 evolved during nitrogen fixation: an uptake hydrogenase that oxidizes the H 2 evolved, and transfers electrons to e.g. the respiratory-chain. As this reaction significantly lowers the H 2 production efficiency of a nitrogenase-based system, targeted mutants with reduced or deficient uptake hydrogenase activity have been produced. This was first achieved by chemical mutagenesis ( Kumar & Kumar, 1991 ; Mikheeva et al. , 1995 ), and later, since the molecular biology tools for genetic engineering were established, by targeted knock-out of structural or accessory genes of the uptake hydrogenase. Uptake hydrogenase-deficient mutants of A. variabilis ( Happe et al. , 2000 ), N. punctiforme ( Lindberg et al. , 2002 , 2004 ), Nostoc sp. PCC 7120 ( Lindblad et al. , 2002 ; Masukawa et al. , 2002 ; Carrasco et al. , 2005 ) and Nostoc sp. PCC 7422 ( Yoshino et al. , 2007 ) have been shown to be significantly better H 2 producers compared with the respective wild types. In general, the H 2 produced by a nitrogenase in the wild type will be quickly reoxidized by the uptake hydrogenase, whereas in an uptake hydrogenase-deficient mutant the H 2 produced will leave the cells. One should bear in mind that all these strains, with the exception of N. punctiforme , also possess a bidirectional hydrogenase. However, only for Nostoc sp. PCC 7120 ( Masukawa et al. , 2002 ) the effect of a hox -defective mutant (Δ hoxH ) has been investigated. A Nostoc sp. PCC 7120 mutant deficient in both hydrogenases (Δ hupL /Δ hoxH ) showed the same increase in H 2 evolution as the uptake hydrogenase-deficient mutant (Δ hupL ), whereas the bidirectional hydrogenase-deficient mutant (Δ hoxH ) produced less H 2 compared with the wild type.
In gas exchange experiments with an uptake hydrogenase-deficient mutant of Nostoc punctiforme ( Lindberg et al. , 2004 ), the amount of H 2 produced per molecule of N 2 fixed varied with the light conditions. The ratio of H 2 produced/N 2 fixed under low light and high light was 1.4 and 6.1, respectively. This showed that, under the specific conditions, the energy flow through the nitrogenase may be directed towards the H 2 production rather than the N 2 fixation.
(2) Low energy efficiency and turnover of the nitrogenase and/or the hydrogenase : H 2 -evolving enzymes with the highest reported turnover are the Fe-hydrogenases ( Houchins et al. , 1984 ; Adams et al. , 1990 ). These enzymes are irreversibly inactivated by oxygen, and are present in e.g. fermentative bacteria (e.g. Clostridium ) and green algae (e.g. Chlamydomonas ) but not in cyanobacteria. An elegant strategy for the creation of an efficient H 2 producer, which will not be inhibited by the surrounding oxygen, would be the expression of a highly active Fe-hydrogenase in the heterocysts of filamentous cyanobacteria unable to reoxidase any H 2 (i.e. an uptake hydrogenase-deficient strain). The heterologous expression of different iron-hydrogenases in various organisms such as Synechococcus ( Asada et al. , 2000 ), E. coli ( Posewitz et al. , 2004 ; King et al. , 2006 ) and Clostridium ( Girbal et al. , 2005 ) has already been achieved. Recently, the accessory genes necessary for the maturation of iron-hydrogenases into active enzymes were identified ( Posewitz et al. , 2004 ; Böck, 2006 ; King et al. , 2006 ). Therefore, the heterologous expression of an active iron-hydrogenase in a cyanobacterial host, e.g. in the heterocyst of a strain for which the genome has been sequenced, is an interesting and realistic project. Moreover, because the iron-hydrogenases are able to use a wide variety of primary electron donors ( Vignais et al. , 2001 ), including ferredoxin, which is the electron donor of the cyanobacterial nitrogenase, it may be possible to link the introduced iron-hydrogenase to an existing electron transfer pathway within the cyanobacterial cell.
(3) Limiting amounts of active H2-evolving enzymes : Because the H 2 -evolving enzymes (nitrogenase(s) and/or bidirectional hydrogenases) are strictly regulated on several different levels (transcription, translation and maturation), one possible way to enhance the production of H 2 may be the overexpression of these enzymes. For this purpose, the genes encoding the selected enzyme/protein to be overexpressed are placed under the control of an artificial promoter and ribosomal-binding site (RBS) combination on an expression vector or placed directly in the genome. The choice of a constitutive or an inducible promoter, together with a strong RBS, takes the enzyme biosynthesis out of the control of the organism's natural regulation system, allowing a significant increase in the amount of enzyme produced.
In heterocystous cyanobacteria grown under N 2 -fixing conditions, c . 5–10% of the vegetative cells differentiate into heterocysts. These specialized cells are compartments with reduced O 2 pressure, and thus suitable for H 2 production, either via nitrogenase or via an introduced hydrogenase. Therefore, the increase of the heterocyst frequency should result in a higher overall H 2 production capacity by the organism. Interestingly, it has been shown that the heterocyst frequency can be increased, e.g. by overexpression of hetR or by inactivation of patS , or hetN ( Buikema & Haselkorn, 2001 ; Golden & Yoon, 2003 ; Borthakur et al. , 2005 ). However, this has not been coupled to H 2 production or increased H 2 production.
(4) High oxygen sensitivity of the nitrogenase and/or the hydrogenase: One main obstacle in H 2 production using photosynthetic microorganisms is the high sensitivity of the H 2 -evolving enzymes, and some attempts have been made to introduce less oxygen sensitive hydrogenases into cyanobacteria. At the Craig Venter Institute (US), work is being carried out aiming at transferring the O 2 -tolerant NiFe-hydrogenase of the purple-sulfur photosynthetic bacterium T. roseopersicina ( Kovacs et al. , 2005 ) into a Synechococcus strain. In addition, several other putative O 2 -tolerant NiFe-hydrogenases have been identified from the marine environment that could be alternative candidates to be introduced into a cyanobacterial background ( Xu et al. , 2005 ). Also in the US, the genes encoding the more O 2 -tolerant NiFe hydrogenase of the purple nonsulfur photosynthetic bacterium Rubrivivax gelatinosus CBS ( Ghirardi et al. , 2005 ), and its accessory proteins, are being introduced into Synechocystis . The in vivo half-life of this hydrogenase is 21 h in air and 6 h in air when the protein is partially purified ( Ghirardi et al. , 2005 ). However, to the authors' knowledge, the heterologous expression of a more oxygen-tolerant hydrogenase in any cyanobacterium remains to be shown.
(5) Electron flow inhibition by accumulation of ATP in a hydrogenase-driven system : In an optimal H 2 production system, all electrons derived from water splitting in PSII should be directed to the H 2 -evolving enzyme (nitrogenase or hydrogenase) to reach maximal energy conversion efficiency. In addition to the electron flow in the photosynthetic electron transport chain, a transmembrane potential is built up that is used for generating ATP through an ATP synthase. In a nitrogenase-based system, the ATP is clearly needed for N 2 fixation. However, in the hydrogenase-catalyzed reaction no ATP is consumed and the electron flow could, in a photosynthetic microorganism, be inhibited by the accumulated transmembrane potential across the thylakoid membrane ( Lee & Greenbaum, 2003 ). It has been observed in the green algae Chlamydomonas reinhardtii that oxygen serves as an electron sink, competing for electrons with the H 2 -producing pathway, resulting in a ‘new oxygen sensitivity’ ( Lee & Greenbaum, 2003 ). A possible solution may be the introduction of a synthetic, polypeptide based on the proton channel into the thylakoid membranes, with its transcription controlled by a hydrogenase promoter ( Lee & Greenbaum, 2003 ). The proton channel may be expressed under H 2 -producing (anaerobic) conditions, to dissipate the proton gradient across the thylakoids membrane. A similar strategy may work for cyanobacteria.
(6) Low quantum efficiency due to too large antennas in both PSII and PSI : In nature, phototrophic organisms have developed to handle and compete during very low light intensities, and, therefore, large antenna systems have been evolved. However, for biotechnological applications in photobioreactors, where an optimal supply with light can be engineered, only a small part of the light will be used by the microorganisms. In addition, self-shading may become a severe limitation causing reduced efficiency in utilizing incoming solar energy. To overcome this problem, a reduction of the antenna size has been proposed ( Melis et al. , 1998 ; Nakajima & Ueda, 1999 ; Lee et al. , 2002 ). The phycocyanin-deficient mutant PD1 of Synechocystis PCC 6714, generated by chemical mutagenesis, showed up to 50% higher maximal photosynthesis activity under high light conditions compared with the wild type ( Nakajima & Ueda, 1997 , 1999 ). Antenna-deficient green algal mutants, created by chemical mutagenesis ( Lee et al. , 2002 ) or genetic engineering ( Polle et al. , 2003 ; Tetali et al. , 2007 ), also showed remarkably greater solar conversion efficiencies and a higher photosynthetic productivity than the respective wild type under mass culture conditions.
(7) Electron-consuming pathways competing with an efficient electron transfer to the H2enzymes : For the production of H 2 through the action of an active nitrogenase or hydrogenase electrons are required to combine with protons to form H 2 . Because protons are abundant within the cell, the main limitation is the available number of electrons. The primary electron donors for the H 2 -producing enzymes are ferredoxin (nitrogenase, Fe-hydrogenases) and NADH/NADPH (NiFe-hydrogenases). However, the electrons are mainly used by other pathways, the so-called ‘competing pathways’, e.g. respiration and the Calvin cycle. Therefore, one strategy for an enhanced H 2 production is to direct the electron flow towards the H 2 -producing enzymes and away from any other competing pathway.
Experiments with the ndhB mutant M55 of Synechocystis PCC 6803, which is defective in the type I NADPH-dehydrogenase complex (NDH-1) ( Cournac et al. , 2004 ), showed that this mutant produces only low amounts of O 2 in the light, has a poor capacity to fix CO 2 and evolves H 2 for several minutes during dark-to-light transitions, while the H 2 uptake was negligible. The electrons used to produce H 2 were mainly coming from water splitting in PSII and from the carbohydrate-mediated reduction of the PQ pool. In another study, the level of reduced NADP shifted from 50% in the wild type to 100% in the NDH-1 mutant ( Cooley & Vermaas, 2001 ). As the cyanobacterial bidirectional hydrogenase evolves H 2 at a relatively high level of reduced NAD(P), the construction of mutants with blocked electron transfer in selected key pathways, increasing the relative level of reduced NAD(P), may be a promising strategy to increase the H 2 production capacity.
Another strategy for directing electrons toward the hydrogenase is to directly link the hydrogenase to PSI. Ihara, (2006b) fused the membrane-bound NiFe hydrogenase from Ralstonia eutropha H16 to the peripheral PSI subunit PsaE of the cyanobacterium Thermosynechococcus elongatus (Hyd-PsaE), and used a PsaE-free PSI (PSI * ) extract from a PsaE-deficient mutant of Synechocystis sp. PCC 6803. The resulting hydrogenase/PSI complex showed light-driven hydrogen production in vitro , which was five times higher compared with a control without a direct coupling of the hydrogenase to PSI. However, as the activity of the hydrogenase-PsaE fusion protein was only 16% of that of the wild-type hydrogenase protein and was totally suppressed by adding ferredoxin (Fd) and ferredoxin-NADP + -reductase (FNR), the authors concluded that the linker between the hydrogenase and PsaE has to be optimized. In another work by the same group ( Ihara et al. , 2006a ), PsaE from Synechocystis sp. PCC 6803 was chemically cross-linked with cytochrome c3 (cytc3) from Desulfovibrio vulgaris and stoichiometrically assembled with PsaE-free PSI to form a cytc3/PSI complex. The NADPH production by this complex coupled with Fd and FNR decreased to c . 10% of the original activity, whereas the H 2 production by the cytc3/PSI complex coupled with hydrogenase from Desulfovibrio vulgaris was enhanced sevenfold. This clearly demonstrated that it is possible, in vitro , to direct the electron-flow toward the hydrogenase by changing the environment of the electron-donating PSI. The next challenge will be to develop an in vivo , or even in situ , functional system based on the assembly of different enzymes and proteins.
As a future perspective, the development of synthetic biology reveals new possibilities for the direct construction of efficient H 2 -evolving cyanobacterial strains. Both in the US (e.g. the Craig Venter Institute) and in Europe (e.g. the EU/NEST project ‘BioModularH 2 ’), the first attempts have been initiated to use this new concept aiming at designing reusable, standardized molecular building blocks that will produce a photosynthetic bacterium containing engineered chemical pathways for competitive, clean and sustainable H 2 production.
Concluding remarks
The fundamental aspects of cyanobacterial hydrogenases, and their more applied potential use as future producers of renewable H 2 from sun and water, are receiving increased international attention. At the same time, significant progress is being made in the understanding of the molecular regulation of the genes encoding both the enzymes as well as the accessory proteins needed for the correct assembly of an active hydrogenase. In the last few years, the transcription factors directly involved in the regulation of cyanobacterial hydrogenases have been identified. Moreover, the first steps to use isolated components from cyanobacteria and other microorganisms in order to create a functional H 2 -producing unit are being taken. With the increasing scientific community and public interest in clean and renewable energy sources, and consequent funding opportunities, rapid progress will be made in the fundamental understanding of the regulation and maturation of cyanobacterial hydrogenases at both genetic and protein levels. Unique and unexpected results in the transcriptional regulation of cyanobacterial hydrogenases will emerge during the coming years. Moreover, the more applied aspects will be highlighted with progress in generating genetically modified strains with an increased capacity for renewable H 2 from sun and water. The possibilities and challenges within synthetic biology, including the use of isolated proteins and parts, will be explored, aiming at creating both cyanobacteria with a high potential for H 2 production as well as functional in vitro systems.
Acknowledgements
This work was financially supported by FCT (POCTI/BIO/44592/2002; SFRH/BD/4912/2001, SFRH/BD/16954/2004), ESF (III Quadro Comunitário de Apoio), the Swedish Research Council, the Swedish Energy Agency, the Nordic Energy Research Program (project BioHydrogen), the EU/NEST Projects SOLAR-H (contract # 516510) and BioModularH 2 (contract # 043340).
References
Editor: Annick Wilmotte